Abstract
Glioblastomas (GBM) can be classified into three major transcriptional subgroups (proneural, mesenchymal, classical), underlying different molecular alterations, prognosis, and response to therapy. However, transcriptional analysis is not routinely feasible and assessment of a simplified method for glioblastoma subclassification is required. We propose an integrated molecular and immunohistochemical approach aimed at identifying GBM subtypes in routine paraffin-embedded material. RNA-sequencing analysis was performed on representative samples (n = 51) by means of a “glioblastoma transcriptional subtypes (GliTS) redux” custom gene signature including a restricted number (n = 90) of upregulated genes validated on the TCGA dataset. With this dataset, immunohistochemical profiles, based on expression of a restricted panel of gene classifiers, were integrated by a machine-learning approach to generate a GliTS based on protein quantification that allowed an efficient GliTS assignment when applied to an extended cohort (n = 197). GliTS redux maintained high levels of correspondence with the original GliTS classification using the TCGA dataset. The machine-learning approach designed an immunohistochemical (IHC)-based classification, whose concordance was 79.5% with the transcriptional- based classification, and reached 90% for the mesenchymal subgroup. Distribution and survival of GliTS were in line with reported data, with the mesenchymal subgroup given the worst prognosis. Notably, the algorithm allowed the identification of cases with comparable probability to be assigned to different GliTS, thus falling within overlapping regions and reflecting an extreme heterogeneous phenotype that mirrors the underlying genetic and biological tumor heterogeneity. Indeed, while mesenchymal and classical subgroups were well segregated, the proneural types frequently showed a mixed proneural/classical phenotype, predicted as proneural by the algorithm, but with comparable probability of being assigned to the classical subtype. These cases, characterized by concomitant high expression of EGFR and proneural biomarkers, showed lower survival. Collectively, these data indicate that a restricted panel of highly sensitive immunohistochemical markers can efficiently predict GliTS with high accuracy and significant association with different clinical outcomes.
Similar content being viewed by others
Introduction
Glioblastoma (GBM), the most frequent and aggressive primary malignant brain tumor in adults, is comprised of a large variety of morphological patterns and histological variants, some of which are included in the recent WHO classification [1]. However, GBM is still considered as a unique entity with standard recognized treatment and tumor heterogeneity reflecting an intrinsic genetic instability. This concept has been recently challenged and it is currently accepted that GBMs can be classified according to their transcriptional profile in three major distinct molecular subgroups (proneural (PN); classical (CL); mesenchymal (MES)), which are associated with specific gene signatures and different prognostic and therapeutic implications [2,3,4]. Expression of genes related to neural (e.g., ASCL1, OLIG2) and glioma-CpG island methylator phenotype [5] defines the PN subtype, frequently associated to a TP53 mutation. Interestingly, the PN subgroup is comprised of IDH1/2 mutated GBMs, and is associated with significantly better prognosis and younger age [6]. In contrast, CL and MES gene signatures are both associated with poor outcome and older age. The CL subtype is characterized by a high frequency of EGFR gene alterations (typically amplification and/or EGFRvIII mutation). Homozygous deletion of 9p21.3 (CDKN2A locus), lack of TP53 mutations, and RB pathways alteration are also frequent [3]. The MES subtype is characterized by high expression of YKL40, MET, and CD44, frequent deletion at 17q11.2 (containing the NF1 gene), chromosomal aberrations in CDK6, CDKN2A, and RB1, and activation of the NF-kB pathway [2]. Interestingly, recurrences are frequently associated with a phenotypic shift into the MES subtype, suggesting that glioma progression may proceed from PN or CL into MES phenotype. Finally, a neural subtype was also described [3], characterized by a gene signature suggestive of a differentiated phenotype with high expression of neuronal markers. However, recent data suggest that this phenotype is nontumor specific but related to contamination of normal neuronal tissue that might also explain lack of distinctive gene abnormalities [2, 7]. Disease stratification may be clinically relevant for identifying potential targets for personalized therapies [8]. We previously reported that stratification of GBM according to EGFR status unmasked a subgroup of EGFR amplified and overexpressing GBM that strongly benefit from metronomic temozolomide-based therapies [9]. Most recently, it was reported that EGFR gene amplification and CL subtypes are associated with significantly shorter time to progression for recurrent GBM treated with bevacizumab [10]. Moreover, we recently showed that the MET receptor kinase, highly expressed in MES subtypes, promotes radio-resistance and is overexpressed in recurring GBMs after radiotherapy, providing preclinical evidences that MET inhibitors can radiosensitize these tumors [11]. Finally, a recent randomized clinical trial showed that addition of bevacizumab to standard radiotherapy plus temozolomide treatment conferred significant OS advantage only in PN GBMs [12]. Overall, these findings indicate that GBM subgrouping carries a significant impact on clinical decision-making aimed to develop subtype-specific therapeutic strategies. However, a feasible standardized approach for subclassification of GBMs in routine diagnostic practice is still lacking. There are few published studies aimed to identify GBM subtypes based on a simplified immunohistochemical (IHC)-based approach [13, 14]. Nevertheless, reported data are neither sensitive nor specific and mostly lack of clinical, molecular, and histopathological correlations. We tried to overcome these limits and, based on an integrated molecular and IHC analysis of a large cohort of GBMs, we propose a simplified IHC-based algorithm able to predict with high accuracy different transcriptional profiles. Finally, yet importantly, we correlated our findings with clinical data, including outcome and response to treatments, identifying GBM subtypes associated to different clinical outcome.
Materials and methods
Patient selection
Newly diagnosed pathologically confirmed GBMs (n = 197) were retrieved from Institutional database of Department of Pathology, Spedali Civili of Brescia. The study was conducted in compliance with the Declaration of Helsinki and policies approved by the Ethical Board of Spedali Civili of Brescia for retrospective and exclusively observational study on archival material obtained for diagnostic purpose for which patient consent was previously collected. Histological diagnosis was revised according to WHO criteria [1] and formalin-fixed paraffin-embedded (FFPE) representative tissue sections for each lesion were selected based on adequate tissue preservation, as assayed by haematoxylin and eosin (H&E) staining. Cases with limited material, such as small stereotactic biopsies, thus not representative of the entire tumor, were excluded. For cases with morphological features suggestive of oligodendroglioma we performed FISH analysis for determination of 1p/19q status. 1p and 19q co-deletion was found only in one case previously diagnosed as GBM, thus excluded from the study. Clinical data were collected from medical records (Table 1). Since patients did not receive uniform treatment we categorized therapeutic schedules, as previously described by Verhaak et al. [3], in “more intensive treatment,” defined as either complete concurrent chemo- and radiotherapy (six cycles) or greater than three subsequent cycles of chemotherapy, and “less intensive treatment,” defined as incomplete concurrent chemo- and radiotherapy (less than six cycles) or less than four cycles of subsequent chemotherapy. A total of 22 patients (15.2%) were not categorized since number of chemotherapy cycles was not available.
Neuropathological and molecular studies
Immunohistochemistry was performed as described in Supplementary Methods. Primary antibodies are listed in Supplementary Table 1. Histopathological features (pleomorphism; presence of gemistocytic, oligodendroglial, small or spindle cell component; epithelioid features; inflammatory infiltrates; glomeruloid vascular proliferation) were semiquantitatively scored as follows: 0, absence; 1, low; 2, moderate; and 3, high. For statistical purposes, values were dichotomized (absence/low, 0; moderate/high, 1). Expression of selected gene classifiers (p53, EGFR, ASCL1, OLIG2, PDGFRα, pNDRG1, YKL40, MET) was semiquantitatively scored on representative tumor regions based on both percentage [score ranges: 0 (0–5%), 1 (6–29%), 2 (30–69%), 3 (≥70%)] and intensity (score ranges: 0, no expression; 1, weak; 2, moderate; 3, high) of immunoreactive (IR) neoplastic cells with a combined cumulative score ranging from 0 to 6. For hierarchical cluster analysis we tailored the cut-off for each marker based on the specific pattern of expression. Indeed, some markers have a baseline expression (e.g., EGFR) and rarely show complete absence of IR cells, while other markers are less widely expressed and rarely reach score 6 (e.g., YKL40). For these markers a score of 3 or 4 has to be considered already related to moderate or strong expression. Therefore, the previous scores (from 0 to 6) were differentially combined using the following scheme: EGFR [0 (0 + 1); 1 (2 + 3 + 4); 2;(5) 3 (6)]; ASCL1, PDGFRα, OLIG2, and p53 [0 (0); 1 (1 + 2); 2 (3 + 4); 3 (5 + 6)]; and YKL40, MET, and pNDRG1 [0 (0); 1 (1 + 2); 2;(3) 3 (4 + 5 + 6)]. The scoring system has been incorporated within the transcriptional status prediction algorithm (available at http://fisher.med.unibs.it:3838/GBMscore) to make it easier to be applied in daily pathology practice. Analysis of O6-methylguanine DNA methyltransferase (MGMT) methylation status, IDH1/IDH2 gene mutations, and FISH analysis for 1p/19q co-deletion and EFGR amplification were also performed. Details in Supplementary Methods.
GBM transcriptional subtypes (GliTS) redux sequencing and data analysis
Transcriptional analysis was performed on 51 representative cases selected from study cohort (n = 197) based on adequacy of material in term of preservation and amount of neoplastic viable cells with exclusion of surrounding normal tissue. RNA was extracted using “High Pure Paraffin kit” (Roche) following the manufacturer’s instructions, quality assessed using RNA-6000 Nano kit on Bioanalyzer-2100 (Agilent), and quantified by RNA-HS Assay kit on Qubit (Life Technologies). Library preparation was performed using a custom TruSeq Targeted RNA kit (Illumina) including 681 probes specific for 280 selected genes: 240 “pre-GliTS redux” genes being the top 30 upregulated and downregulated genes according to the Verhaak’s study where a 840 gene signature defining four GliTS was identified [3]; 25 additional cancer genes; and 15 housekeepers (Supplementary Table 2). Based on percentage of RNA fragments longer than 200 nucleotides (DV200) ≥ 400 ng of total RNA were used for library preparation, following the manufacturer’s indications (Illumina). Quality and molarity of libraries were calculated using DNA-HS assay kit on Bioanalyzer-2100 (Agilent) on Qubit. Libraries were pooled and sequenced on a Miseq (Illumina). Sequences were mapped on GRCh38 using splice aware mapper MapSplice [15] and read counts were calculated with HTseq [16], gene annotation performed on gencode 22 [17]. Globally we could successfully map 267 unique gene symbols. After validating that ssGSEA robustly classifies TCGA samples using the limited number of genes selected as above (data not shown), classification of GBM samples was performed with ssGSEA implementation provided in Wang et al. [2] according to the restricted CL, PN, and MES subtypes upregulated signature genes “GliTS redux” included in our profile and validated on TCGA dataset. For each patient we generated CL, PN, and MES specific metagenes (i.e., aggregate patterns of gene expression) by evaluating the average of L2R expression values of subtype-specific GliTS redux genes.
Cluster analysis and transcriptional prediction algorithm
Hierarchical cluster analysis was performed using Kendall correlation coefficient as similarity metric and Ward criterion [18]. Transcriptional status prediction was performed using random forest algorithm [19] growing 500 trees with two predictors sampled for splitting at each node. Subtype assignment probability was computed on OOB (out-of-bag) samples. A pairwise proximity matrix was computed from random forest on OOB samples, based on the frequency that pairs of data points end up in. Multidimensional scaling was used to visualize sample similarities on a Cartesian plane. Transcriptional status prediction analysis was performed based on combination of IHC and transcriptional data applying the algorithm previously described. In order to obtain the best combination of biomarkers able to predict the transcriptional profile itself and improve performance of the transcriptional status prediction algorithm, we selected only cases with either a strong assignment of GliTS redux subtype or an unequivocal IHC profile. The final transcriptional status prediction analysis was performed on 39 cases out of 51 previously submitted to transcriptional analysis. We included 37 GBM strongly assigned to the specific subgroup, as assessed by high significant p-value. Four cases were excluded because not significantly associated to any GliTS redux subtype. For samples not univocally associated to a specific GliTS redux subtype we considered the value of the metagenes correlated to the subtypes and we included in the analysis only cases with a robust association to a single subgroup (differential expression of at least >0.50). Cases with an extremely heterogeneous IHC profile according to the expression of gene classifiers were also excluded. We finally included two additional cases from the studied cohort with the R132H-IDH1 mutation, considered by default as PN. OOB probability of subtype’s membership were computed from random forest model for all samples and used to rank them for display.
Statistical analysis
Clinical, pathological, and molecular variables were dichotomized for statistical purposes using cut-off values. Correlations among biomarkers were computed using Spearman’s rank method. Descriptive statistics were used for patient cohort and survival calculated by Kaplan–Meier method and compared with log-rank test. Clinical, pathological, and molecular variables were considered for interaction in the Cox proportional-hazards model. Both univariate testing and multivariate analysis were used and Bonferroni correction applied to correct for multiple testing. REMARKS guidelines were followed. For Student’s t test analysis p-value < 0.05 was considered statistically significant. We used SPSS software version 20.0. Details in Supplementary Methods.
Results
Patient cohort and clinical-pathological features
The study was conducted on 197 representative newly diagnosed GBMs upon revision of histological diagnosis. Clinical information with complete follow-up was available for 192 cases (97.5%). Median age at diagnosis was 60.8 years (range: 25–83) and female/male ratio was 1:1.8 (female = 70, male = 127). Clinical features, including KPS and recursive partitioning analysis (RPA) classification are summarized in Table 1. All patients underwent partial or subtotal tumor resection and only three patients were subjected to open-surgery biopsy. In 62.9% of patients, surgery resulted in complete removal with no residual enhancement seen on postoperative CT or MRI scans. Partial removal occurred in 70 patients (35.5%). Only ten patients (5.1%) did not receive any therapy due to sudden worsening of clinical conditions. A total of 37 patients (18.8%) received radiotherapy or chemotherapy alone, whereas radiotherapy and concomitant and sequential temozolomide was the most frequently adopted chemotherapeutic schedule (73.6%). Since patients did not receive uniform treatment we categorized therapeutic schedules in “more intensive treatment” and “less intensive treatment,” as previously described by Verhaak et al. [3].
A simplified transcriptional gene signature allows identification of GliTS
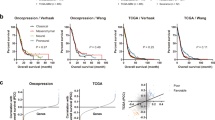
Transcriptome analysis to assign GliTS was carried out on 51 FFPE tissue samples from study cohort (25.9%) based on representativeness of neoplastic areas and adequate tissue preservation. Classification was performed according to the original algorithm, published by Wang et al. [2], adapted for GliTS redux (see Materials and methods and Fig. 1a). Notably, when applied to the cohort of tumors originally employed for clinical and molecular validation of GliTS (TCGA, n = 497 primary tumors included irrespectively of their IDH status, data retrieved from http://gliovis.bioinfo.cnio.es/) [20], GliTS redux maintained a high level of correspondence with the original GliTS [concordant samples 89.34%, Cohen’s kappa = 0.84, (0.8, 0.88)] (Fig. 1b; http://gliovis.bioinfo.cnio.es/). GliTS redux was successfully applied to our cohort and identified 17 CL (40.48%), 12 PN (28.57%), and 13 MES (30.96%) patients, which were univocally assigned to their class with proportions similar to the reference dataset (37.62% CL, 30.95% PN, 30.18% MES) (Fig. 1c). Five samples showed enrichment of multiple classes and four patients were not assigned (Supplementary Table 3). This observation was consistent with a previous report showing that a subset of TCGA samples cannot be univocally associated with a single GliTS subtype due to intrinsic GBM heterogeneity, as determined by single-cell sequencing [2].
a Flow-chart of methodology used for RNAseq profiling and classification performed according to the original algorithm, published by Wang et al. [2] and adapted for GliTS redux. b Caleydo view (top) of correspondences between the GliTS and GliTS redux class assignments, representing the numbers of samples classified by the two signatures as reported in the table (bottom). Our proposed “GliTS redux” maintained a high level of correspondence with the original “GliTS” generated on the TGCA reference cohort [Cohen's kappa = 0.84, (0.8, 0.88)]. c Heatmap of “GliTS redux” genes sorted by gene expression subtype in study cohort. When applied to our study cohort “GliTS redux” allowed to successfully identify GBM subgroups.
A restricted panel of selected biomarkers discriminates between different GBMs
We then investigated if a simplified IHC-based approach could be suitable to adequately identify different GliTS. To this end, selection of appropriate gene classifiers was crucial. Biomarkers were selected from the same gene set used for identification of transcriptional profile. Gene classifiers were differentially expressed and, most importantly, inversely correlated among different GBMs, allowing for discrimination and clustering (Fig. 2a). We first chose to investigate EGFR expression as a strong predictive biomarker of CL. EGFR amplification and overexpression are strongly associated with CL GBM gene signature and we have previously reported that EGFR overexpression is strictly related to EGFR gene amplification [9]. FISH analysis performed on 106/197 (53.8%) cases confirmed that EGFR-overexpressing GBMs (cumulative score = 6) were mostly amplified (46/49; 93.9%). Interestingly, among nonamplified cases (n = 36) we found an EGFR cumulative score = 6 only in three cases (3/36; 8.3%). In addition, as also reported [3], EGFR overexpression resulted inversely correlated with p53 (correlation index −0.17) and MES biomarkers (YKL40, MET, pNDRG1; correlation index −0.29, −0.23, −0.29, respectively) (Fig. 2a). Likewise, we recently reported that NDRG1, whose expression is significantly inhibited by ASCL1, a robust gene classifier for PN [3, 21], can be considered a novel gene classifier of MES GliTS [22]. Data confirmed that expression of pNDRG1 and ASCL1 were inversely correlated (correlation index −0.26) and the former was expressed together with other MES specifiers, YKL40 and MET (correlation index +0.60 and +0.35, respectively; Fig. 2a). Interestingly, we previously reported that MET expression is associated with glioma stem-like cells with a MES or PN gene signature, but virtually absent and mutually exclusive in EGFR expressing and amplified glioma stem-like cells with CL profile [23]. Accordingly, in our cohort MET expression was mostly mutually exclusive with EGFR (correlation index −0.23). We then selected PDGFRα, OLIG2, and ASCL1 as biomarkers related to PN subgroup [3]. Of note, PN gene classifiers were found to be mostly inversely correlated with MES biomarkers (Fig. 2a) and highly expressed in IDH1-mutated GBMs (n = 3), considered by default as PN [3, 5]. Overall, these data confirm that selected biomarkers are sensitive and specific and their combined expression profiles allow discriminating between different GBMs.
a Immunostaining of gene classifiers on representative cases (left panel) and correlation index between their expression (right panel). As shown, selected biomarkers were found to be mostly inversely correlated and differentially expressed between different GBMs, allowing for discrimination and clustering. Images are from ×20 original magnification. b Dendrogram from cluster analysis based on combination of biomarkers expression shows distribution of GBMs in three major branches and their relative expression profiles within single group (plots). The expression profile allows to identify different tumor expression profiles highly expressing PDGFRα-OLIG2-ASCL1, EGFR, and YKL40-MET-pNDRG1, respectively. The lower string shows GliTS assigned by the machine-learning algorithm (available at http://fisher.med.unibs.it:3838/GBMscore) to all 197 GBMs and based on combination of IHC and transcriptional data. Pie charts show distribution of predicted GliTS within the three branches previously identified within the dendrogram. Comparison between cluster analysis and transcriptional status prediction allowed to identify an additional GBM subgroup (PN2) with concomitant high expression of EGFR and PN metagenes. c Clustering based on prediction proximity (upper panel) and probability of prediction (lower panel) indicate that MES and CL subgroups are well segregated, while PN GBMs frequently share overlapping features between other subgroups, with large majority of nonconcordant cases related to a mixed PN/CL profile.
A machine-learning transcriptional status prediction algorithm efficiently assigns GliTSs
We first performed a hierarchical cluster analysis based on combination of the expression of the selected gene classifiers. Analysis identified different branches within the dendrogram that could be potentially assigned to a specific transcriptional profile according to high or low level of expression of the different gene classifiers (Fig. 2b). Interestingly, the clustering dendrogram showed well segregated branches identifying: (i) a group of tumors enriched in PDGFRα, OLIG2, and ASCL1 expression, associated to PN GliTS; (ii) a group of tumors highly expressing EGFR, associated to CL GliTS; and (iii) a group of tumors highly enriched in YKL40, MET, pNDRG1, and p53, associated to MES GliTS. Indeed, attribution of the specific subgroup remains arbitrary. To overcome this problem, we performed a transcriptional status prediction analysis based on combination of IHC and transcriptional data applying a specific algorithm in order to obtain the best combination of biomarkers able to predict the transcriptional profile itself. GBMs (n = 39), selected based on either a strong assignment of GliTS redux subtype and an unequivocal IHC profile as previously described, were subjected to transcriptional status prediction analysis by comparing their IHC and transcriptional profiles. The algorithm (available at http://fisher.med.unibs.it:3838/GBMscore) assigned a specific subgroup to all 197 GBMs, independently from presence or absence of transcriptional data and based only on IHC scores (Fig. 2b). Predicted GliTS showed the following distribution: PN 84/197 (42.6%); MES 62/197 (31.5%); and CL 51/197 (25.9%). When analyzed separately, concordance between profiles predicted by the machine-learning algorithm and corresponding transcriptional signatures (available for 39 cases) was 79.5% (31/39). Interestingly, concordance for CL and MES subgroups was 81.3% (13/16) and 90% (9/10), respectively, dropping down to 69.2% (9/13) for the PN subgroup. Considering clustering based on prediction proximity it can be noted that 19% (3/16) among CL samples are clustered to other subtypes (two MES, one PN) and 31% (4/13) PN are more similar to other subtypes, mostly CL (three CL, one MES). Conversely, all MES are clustered together (Fig. 2c). Thus, these data indicate that MES and CL subgroups were well segregated, while PN GBM more frequently shared overlapping features between the groups. Of note, the large majority of nonconcordant cases (7/8) were related to a mixed PN/CL (5/8) or PN/MES (2/8) profiles. Actually, the machine-learning algorithm assigned the GliTS to all cases, but it has to be noted that some cases had comparable probability to be assigned to at least two GliTS, thus falling within overlapping regions, as shown in Fig. 2c. Data reflect the huge heterogeneous phenotype of some GBMs, particularly with PN phenotype. Indeed, the algorithm imputes a probability for each GBM to be assigned to a specific subgroup, thus highlighting cases with high tumor heterogeneity. Interestingly, combination between cluster analysis and transcriptional status prediction identified a GBM with a peculiar profile, assigned to PN by the prediction algorithm, but that fell within the CL branch of the dendrogram. These samples, characterized by concomitant high expression of EGFR and PN gene classifiers, will be from now identified as PN2 (n = 30), while remaining PN cases, with robust PN profile and lower EGFR expression, will be identified as PN1 (n = 26) (Fig. 2b and Supplementary Fig. 1).
Different morphological features are associated to different GliTS
Morphological features of all GBMs were correlated to predicted GliTS, also considering distinction between PN1 and PN2. Of note, 40.1% of patients showed GBMs with small cell morphology, a feature significantly associated to CL subgroup (Fig. 3a) [24]. Dense vascular network and glomeruloid features, reflecting intense tumor neo-angiogenesis, were also associated to CL (Fig. 3b). Interestingly, both features were significantly present in PN2 GliTS, which closely recapitulates morphological features of CL. As expected, albeit only present in 12.2% of cases, oligodendroglial features were found predominantly within PN1 GliTS (Fig. 3c). Conversely, gemistocytes, spindle cells, pleomorphism, and epithelioid features (Fig. 3d–g) were significantly associated to MES GliTS and barely detectable within other subgroups, with the exception of PN1, that exhibits a variable degree of pleomorphism and gemistocytes. Of note, inflammatory component was significantly associated to MES GliTS (Fig. 3h), as recently described [2].
GliTS classification identifies clinically relevant GBM subtypes
Survival information was obtained for all patients (100%), while complete clinical data were available for 192 cases (97.5%). Indeed, we did not include in survival analysis IDH1-mutant GBMs (n = 3; median OS 35.3 months), all predicted as PN1 by the algorithm and considered as a separate entity with favorable outcome [5, 7]. When comparing outcomes among the predicted transcriptional subgroups on the remaining 189 patients (Fig. 4a), we observed a better survival rate in CL GBMs as compared with PN and MES (CL vs PN vs MES: median OS 19.4 vs 12.0 vs 11.2 months; p = 0.003 by log-rank test) (Fig. 4b). Univariate Cox-regression analysis on the same cohort of patients indicated that younger age, higher KPS, III or IV Radiation Therapy Oncology Group RPA (RTOG-RPA) classes, MGMT promoter methylation and concomitant radio–chemotherapy, and/or sequential temozolomide treatment were all associated with significant better survival (Table 2). Interestingly, among the different gene classifiers analyzed, only YKL40 and MET overexpression were significantly correlated to worse prognosis (Supplementary Table 4), as also reported [11]. As expected, YKL40 and MET overexpressing cases were predicted as MES. At multivariate Cox-regression analysis RTOG-RPA classes, MGMT promoter methylation and radio–chemotherapy treatment remained statistically significant (Table 2). After Bonferroni correction for OS only MGMT promoter methylation and radio–chemotherapy treatment remained statistically significant (p = 0.01). In our cohort, therapy persisted as favorable prognostic factor across different predicted subgroups with better survival for patients receiving combined radio–chemotherapy treatment as compared with palliative radiotherapy or chemotherapy only (median OS: CL 20.5 vs 9.5 months; PN 15.4 vs 7.1 months; MES 14.6 vs 5.4 months). Interestingly, within MES subgroup, patients receiving radio–chemotherapy represented only 60.3% (38/63), while CL and PN were 86% (43/50) and 81.2% (65/80), respectively, suggesting that MES GBMs show compromised clinical conditions already at first clinical presentation that impinge the gold standard treatment. Since therapeutic schedules resulted the variable that mostly affects survival, we examined the efficacy of more (n = 93) or less (n = 30) intensive radio–chemotherapy treatment, defined as previously described, excluding patients that did not received any therapy (n = 10) or only palliative treatment (n = 37) (Fig. 4a). Again, outcomes of predicted GliTS (n = 123) showed better OS for CL as compared with PN and MES (CL vs PN vs MES: median OS 21.0 vs 14.3 vs 15.0 months), albeit not reaching statistical significance (overall comparison p = 0.166 by log-rank test) (data not shown). At univariate Cox-regression analysis both MGMT promoter methylation, more intensive treatment and younger age resulted statistically significant, while at multivariate Cox-regression analysis only more intensive treatment remained significant, also after Bonferroni correction for OS. None of the gene classifiers was significantly associated with survival in this cohort (Supplementary Table 4). We further stratified patients according to their subtype and more or less intensive radio–chemotherapy treatment. While aggressive treatment strongly improved survival in CL (median OS for less vs more intensive treatment, 8.4 vs 23.1 months; p < 0.0005 by log-rank test), it did not significantly alter OS in both PN and MES (median OS for less vs more intensive treatment: 12.9 vs 16.5 for PN and 14.8 vs 15.4 months for MES; p = 0.084 and p = 0.525 by log-rank test, respectively) (Fig. 4c). Of note, PN showed an unexpectedly low OS comparable with MES GBMs. Since cluster analysis allowed to highlight two different PN profiles, PN1 and PN2, we performed OS analysis including these categories in patients receiving more intensive treatment (n = 93). Interestingly, PN2 showed the poorest prognosis, even worse than MES GBMs (median OS for CL vs PN1 vs MES vs PN2, 23.1 vs 20.1 vs 15.4 vs 12.0 months; overall comparison p = 0.016 by log-rank test) (Fig. 4d). In addition, MGMT methylation status (available for 103/123; 83.7%) resulted predictive to temozolomide responsiveness only in CL (median OS for NM vs M, 19.7 vs 25.9 months; p = 0.025 by log-rank test), but not in PN and MES subtypes. Interestingly, PN2 is comprised of a small percentage of methylated cases as compared with PN1 (PN1 = 42.5%, PN2 = 20.0%). PN1/PN2 clinical features are summarized in Supplementary Table 5.
a Flow-chart showing exclusion criteria for survival analysis. b Kaplan–Meier estimates of survival between different GliTS (n = 189). Data show a better survival rate in CL GBMs as compared with PN and MES (p = 0.003 by log-rank test). c Since therapy was found both at uni- and multivariate Cox-regression analysis as a favorable prognostic factor across different GliTS we selected only patients receiving combined radio–chemotherapy treatment (n = 123), further stratified according to more (n = 93) or less (n = 30) intensive radio–chemotherapy treatment. More intensive treatment significantly improved survival only in CL (p < 0.0005 by log-rank test). d Since PN showed an unexpectedly low OS we further stratified PN GliTS in PN1 and PN2. Survival analysis for patients receiving more intensive treatment (n = 93) for CL, PN1, PN2, and MES subgroups showed that PN2 had the poorest prognosis (p = 0.016 by log-rank test). CI confidence interval; HR hazard ratio.
Discussion
Classification based on transcriptional gene signature has emerged as a valuable tool to investigate GBM biology and enable uniform reliability in reporting scientific data. Transcriptional signatures were associated with different molecular alterations [3], key signaling pathways [25], and DNA methylation [5]. Thus, a specific transcriptional profile potentially relates to different gene classifiers expression. However, there are few studies aimed to correlate biomarkers expression to transcriptional signatures [13, 14] and no public datasets are available directly correlating transcriptional and immunophenotypical profiles within the same patient cohort. In a previous report, the attempt to provide an IHC-based approach was validated at the transcriptional level only in a separate cohort of patients [14]. Moreover, GliTS classification has not yet become part of widespread clinical practice mainly due to lack of reliable clinical laboratory assays to measure RNA expression from FFPE samples. We tried to overcome these limits by a simplified method to assign GliTS to GBM on routine FFPE samples. We first developed a novel “GliTS redux” approach according to the original algorithm published by Wang et al. [2], adapted for a restricted gene signature that was found to be highly robust, as indicated by high correspondence upon validation on the TCGA dataset. GliTS redux was successfully applied, univocally assigning specific subgroups with similar proportion to the reference dataset. Interestingly, a subset of samples (17.6%) was not significantly or not univocally associated to a specific GliTS, in line with previous reports and ascribed to intrinsic tumor heterogeneity, as determined by single-cell sequencing [2, 26]. It was recently reported that a single GBM comprises tumor cells at different developmental cellular states associated to different phenotypes [27]. Of note, authors suggest that the relative frequency of each cellular state is influenced by underlined genetic and epigenetic alterations and varies between different GBM samples. Thus, association between molecular alterations and cellular states could explain tumor heterogeneity with the more frequent cellular state having the higher impact on the expression profile of tumor bulk and attribution to a specific transcriptional subtype. Indeed, there are obvious limitations to molecular methods commonly used for expression profiling. Analysis based on homogenized tissue samples may be not completely representative of the original tumor due to contamination of microenvironment that may represent up to 70% of tumor bulk [28] and could obscure the real transcriptional signature of neoplastic cells, as we previously reported [29]. Of note, a rich inflammatory component is strongly associated to poor prognosis and contributes to the MES gene signature [2]. Accordingly, we show that inflammatory component and worst prognosis are significantly associated to predictive MES GliTS. Thus, distinctive GliTS and clinical features can be ascribed, at least in some cases, to abundant tumor-associated microenvironment. In addition, as described, a single tumor may activate more than one transcriptional profile contributing to increased transcriptional heterogeneity [2, 27], as also suggested by surgical multisampling from the same patient showing that some lesions display different GBM subtypes within the same tumor [30]. We also recently showed that a phylogenetically related glioma stem-like cell hierarchy can be found in matched primary and recurrent GBMs reflecting genetic evolution under therapeutic pressure and tumor heterogeneity [31]. This temporal and spatial heterogeneity would render transcriptional classification relatively challenging and not necessarily representative of the tumor as a whole. Histopathological and immunophenotypical analysis allows to recognize different morphological aspects within the entire resected sample that may reflect this heterogeneity. Indeed, the machine-learning algorithm allowed assigning a probability score for each GBM to belong to a specific GliTS, with some cases having comparable probabilities to be assigned to different GliTS, reflecting their heterogeneous phenotype. These observations suggest that established GBM gene classifiers may be variably expressed across individual cells within the same tumor and an IHC-based approach may help to unmask the real tumor expression profile since biomarkers are scored only in neoplastic cells, highlighting their distinct distribution within different tumor components. Moreover, transcriptional analysis, albeit simplified, is still an expensive and complex procedure not fully available to all neuropathology services. The proposed IHC-based approach, using a restricted panel of well-recognized subgroup-specific gene classifiers, is simple and reproducible. Biomarkers were essentially mutually exclusive or inversely correlated, features extremely useful to perform a cluster analysis sufficiently sensitive and specific for discerning different GliTS. Reported data show high concordance between profiles obtained either by molecular or IHC-based approach, being 81.3% for CL and up to 90% for MES. Interestingly, concordance dropped down to 69.2% for PN. Clustering based on prediction proximity confirmed that CL and, particularly, MES subtypes were consistently clustered separately, while PN frequently shared overlapping features with other GliTS, reflecting a heterogeneous phenotype. Of note, when analyzed together IHC-based hierarchical cluster analysis and transcriptional status prediction identified cases with a hybrid mixed PN/CL phenotype (PN2), assigned to PN subgroup by prediction algorithm, but falling within CL branch of the IHC-based cluster dendrogram. Indeed, PN2 cases have been predicted by the algorithm as PN, but with comparable probability to be assigned to the CL subgroup, mainly due to the high expression of EGFR. Of note, survival analysis showed that PN GliTS has an unexpectedly low OS, but when analyzed separately only PN2 showed poor prognosis. As shown, PN2 has concomitant high expression of EGFR and PN biomarkers. Indeed, we previously reported that EGFR expression identifies functionally and molecularly distinct population of tumor-initiating stem-like cells and is required for gliomagenesis [32]. We can speculate that sustained EGFR expression in PN2 may be related to an immature phenotype and combination of PN phenotype with EGFR expression confers to neoplastic cells an aggressive behavior. Interestingly, the original report from Verhaak et al. [3] showed that PN GliTS did not have any benefit from more intensive therapeutic schedules, as opposed to other subtypes. Accordingly, our data show that aggressive treatment strongly improved survival only in CL and not in PN. In addition, an EGFR-dependent NF-κB activation has been reported in GBM [33], playing a key role in MES differentiation [34] and promoting PN-to-MES transition, either spontaneously or in response to therapy [35, 36]. We can thus speculate that PN2 GliTS may activate alternative molecular pathways closely related to MES phenotype leading to aggressive behavior and temozolomide resistance. Contrariwise, EGFR upregulation in CL GliTS is mediated by molecular pathways that allow better response to intensive temozolomide treatment with improve survival, as we previously suggested [9] and also reported hereby. In addition, PN2 GliTS is comprised of a small percentage of MGMT methylated cases, feature that conversely characterize PN1 and correlated to temozolomide responsiveness. To date, translational significance of GliTS classification has been impinged by lack of effective specific subtype therapies. GliTS classification combined with an IHC-based approach could help in identifying biomarkers differentially expressed within specific GliTS that may help identifying druggable molecules for personalized therapies. Actually, a recent study suggests that IDH1 wild-type PN GBMs benefit from first-line bevacizumab treatment [12]. We also reported that MES differentiation promotes radiation resistance and GBMs may be radiosensitized by MET pharmacological inhibition, robustly expressed in MES GliTS [11, 37]. Subgrouping also provides insights into GBM biology and histogenesis. Different transcriptional profiles may be associated with distinct mechanisms of gliomagenesis. However, mechanisms of subgroup specification are currently unknown. Indeed, we recently reported that enforced expression of ASCL1, a robust PN biomarker, promotes acquisition of the PN phenotype and concomitantly represses MES features by direct downregulation of NDRG1 expression. Conversely, either inhibition of ASCL1 or enforced NDRG1 expression in PN glioma stem-like cells induce PN-to-MES transition [22]. Finally, we also highlight histological features significantly correlated to different GliTS, an observation completely absent in the literature and useful in routine diagnostic procedures. In summary, we developed a novel transcriptional status prediction algorithm (http://fisher.med.unibs.it:3838/GBMscore) based on combination of transcriptional and IHC data, easily applicable in daily clinical practice, that assigned GliTS to all cases in an objective manner. We are aware that reliability of the approach could be further improved by future identification of sensitive and specific gene classifiers and by increasing the number of cases to be submitted to the predictive algorithm. However, we think that our results improve understanding of GBM subtype classification, elucidate the critical correlation between tumor heterogeneity and immunophenotype, and may provide a useful tool to stratify patients in clinical trials for personalized therapies.
References
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–20.
Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33:152.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110.
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73.
Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22.
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77.
Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232:165–77.
Cominelli M, Grisanti S, Mazzoleni S, Branca C, Buttolo L, Furlan D, et al. EGFR amplified and overexpressing glioblastomas and association with better response to adjuvant metronomic temozolomide. J Natl Cancer Inst. 2015;107:djv041.
Hovinga KE, McCrea HJ, Brennan C, Huse J, Zheng J, Esquenazi Y, et al. EGFR amplification and classical subtype are associated with a poor response to bevacizumab in recurrent glioblastoma. J Neurooncol. 2019;142:337–45.
De Bacco F, D’Ambrosio A, Casanova E, Orzan F, Neggia R, Albano R, et al. MET inhibition overcomes radiation resistance of glioblastoma stem-like cells. EMBO Mol Med. 2016;8:550–68.
Sandmann T, Bourgon R, Garcia J, Li C, Cloughesy T, Chinot OL, et al. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: retrospective analysis of the AVAglio trial. J Clin Oncol. 2015;33:2735–44.
Le Mercier M, Hastir D, Moles Lopez X, De Neve N, Maris C, Trepant AL, et al. A simplified approach for the molecular classification of glioblastomas. PLoS ONE. 2012;7:e45475.
Conroy S, Kruyt FA, Joseph JV, Balasubramaniyan V, Bhat KP, Wagemakers M, et al. Subclassification of newly diagnosed glioblastomas through an immunohistochemical approach. PLoS ONE. 2014;9:e115687.
Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9.
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74.
Murtagh F, Legendre P. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J Classif. 2014;31:274–95.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19:139–41.
Park NI, Guilhamon P, Desai K, McAdam RF, Langille E, O’Connor M, et al. ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell. 2017;21:411.
Narayanan A, Gagliardi F, Gallotti AL, Mazzoleni S, Cominelli M, Fagnocchi L, et al. The proneural gene ASCL1 governs the transcriptional subgroup affiliation in glioblastoma stem cells by directly repressing the mesenchymal gene NDRG1. Cell Death Differ. 2018;26:1813–31.
De Bacco F, Casanova E, Medico E, Pellegatta S, Orzan F, Albano R, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Stem Cells. 2012;72:4537–50.
Perry A, Aldape KD, George DH, Burger PC. Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer. 2004;101:2318–26.
Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE. 2009;4:e7752.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–401.
Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–49.e21.
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–9.
Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–14.
Orzan F, De Bacco F, Crisafulli G, Pellegatta S, Mussolin B, Siravegna G, et al. Genetic evolution of glioblastoma stem-like cells from primary to recurrent tumor. Stem Cells. 2017;35:2218–28.
Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–13.
Yang W, Xia Y, Cao Y, Zheng Y, Bu W, Zhang L, et al. EGFR-induced and PKCepsilon monoubiquitylation-dependent NF-kappaB activation upregulates PKM2 expression and promotes tumorigenesis. Mol Cell. 2012;48:771–84.
Yamini B. NF-kappaB, mesenchymal differentiation and glioblastoma. Cells. 2018;7:125.
Segerman A, Niklasson M, Haglund C, Bergstrom T, Jarvius M, Xie Y, et al. Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17:2994–3009.
Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300.
De Bacco F, Luraghi P, Medico E, Reato G, Girolami F, Perera T, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–61.
Acknowledgements
We thank Sara Cancelli, Elisabetta Spada, and Elisabetta Sandrini for collecting data and technical support, Gigliola Reato and Daniela Cantarella for technical support with RNAseq profiles, and James Hughes for revising the manuscript. A special thanks goes to Salvatore Grisanti for his suggestions and helpful critical discussions. Work was conducted on behalf of Neuro-Oncology Group, Spedali Civili, University of Brescia.
Funding
This work was supported by Italian Ministry of Health (Grant RF-2016-02361014) and “Associazione dedicato A te” to PLP; Italian Association for Cancer Research (AIRC; Investigator Grant no. 19933) and “Comitato per Albi 98” to CB; Italian Association for Cancer Research (AIRC; Investigator Grant IG16823) to RG; Italian Ministry of University (PRIN; projects no. 20 178S4EK9) to SC.
Author information
Authors and Affiliations
Consortia
Contributions
Conceptual design and critical revision of data: PLP, CB, and RG; bioinformatic analyses: SC, CI, and EM; RNAseq and molecular analysis: FO, CI, FP, and FDB; methodological expertise and performed most of the experiments: FO, FP, MC, and FDB; clinical data revision and sample collection: LT, MB, MMF, PPP, and RL; neuropathology, IHC, FISH, and revision of clinical diagnosis: PLP, FP, MC, DM, and PB; and manuscript preparation and critical revision: PLP, CB, RG, FO, FP, and MC. All authors contributed to data analysis/interpretation and approved the final version of this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
All procedures was conducted in compliance with Declaration of Helsinki and policies approved by Ethical Board of Spedali Civili of Brescia for retrospective and exclusively observational study on archival material obtained for diagnostic purpose for which patient consent was previously collected. Clinical information were obtained on behalf of Neuro-Oncology Group, Spedali Civili, University of Brescia. Article does not contain any studies with animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Orzan, F., Pagani, F., Cominelli, M. et al. A simplified integrated molecular and immunohistochemistry-based algorithm allows high accuracy prediction of glioblastoma transcriptional subtypes. Lab Invest 100, 1330–1344 (2020). https://doi.org/10.1038/s41374-020-0437-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-020-0437-0
This article is cited by
-
Necrotic reshaping of the glioma microenvironment drives disease progression
Acta Neuropathologica (2022)
-
Molecular Subgroup Classification in Alzheimer’s Disease by Transcriptomic Profiles
Journal of Molecular Neuroscience (2022)
-
Simulating the behaviour of glioblastoma multiforme based on patient MRI during treatments
Journal of Mathematical Biology (2022)