Abstract
The mechanisms underlying the lineage switching from prostate adenocarcinoma (AdPC) to lethal neuroendocrine prostate cancer (NEPC) have yet to be completely elucidated. In this study, RNA sequencing data from a unique patient-derived xenograft NEPC model and a clinical NEPC cohort were used to identify the potential genes driving NEPC progression. Enrichr analysis resulted in the identification of SRY-related HMG-box gene 2 (SOX2) as a potential repressor that causes decrease in the expression of AdPC specific genes in NEPC. Assays involving the stable overexpression of SOX2 in LNCaP and CWR22RV1 cells validated this role of SOX2 in vitro. Mechanistic studies showed that the repressor role of SOX2 was attributed to the marked global hypomethylation of histone H3, which was driven by the activation of lysine-specific demethylase 1 (LSD1). Furthermore, Enrichr also predicted SOX2 as a driver gene involved in the upregulation of NEPC specific genes. However, SOX2 alone could only marginally induce the expression of some neuroendocrine markers in vitro, which was consistent with previous reports. Moreover, we also elucidated the molecular features of LNCaP-SOX2 cells that may confer resistance to androgen-deprivation therapy (ADT) and the inclination toward neuroendocrine transdifferentiation. The results of this study reveal a novel mechanism for SOX2 in the progression of NEPC via LSD1-mediated global epigenetic modulation. This discovery suggests that LSD1 may be a selective target for the prevention of NEPC progression.
Similar content being viewed by others
Introduction
Androgen-deprivation therapy (ADT) is the primary strategy used for patients with advanced prostate cancer, but despite its initial effectiveness, ADT inevitably leads to the development of castration-resistant prostate cancer (CRPC) [1,2,3]. The most recognized androgen receptor (AR) negative CRPC variant is neuroendocrine prostate cancer (NEPC), which is highly aggressive and has a poor prognosis.
The transition from prostate adenocarcinoma (AdPC) to neuroendocrine prostate cancer (NEPC) is molecularly characterized by the acquisition of neuroendocrine (NE) phenotypes and a decrease/loss of the adenocarcinoma luminal phenotype [4,5,6,7]. Many important alterations at the genetic, transcriptional, and epigenomic levels that drive the transition from AdPC to NEPC have been discovered recently. The co-overexpression and gene amplification of MYCN and AURKA, which work in synergy to promote the aggressiveness and the development of the NE phenotype in AdPC, are present in a subset of NEPC tumors [6, 8, 9]. RB1 and TP53 are commonly concomitantly mutated in ADT-induced NEPC variants [10,11,12,13]. However, RB1 or TP53 depletion are unlikely to play a direct role in NEPC transdifferentiation but rather facilitate prostate cancer lineage plasticity, which is increasingly appreciated as a potential mechanism that predisposes AdPC cells to stably establish an NEPC phenotype once they transdifferentiate [14, 15]. In this NEPC predisposing genetic context, treatment with potent AR pathway inhibitors (such as enzalutamide and abiraterone) release the previously AR-repressed NE master regulators, such as SRY-related HMG-box gene 2 (SOX2) [16], BRN2/POU3F2 [17], MYCN [8, 18], SRRM4 [15, 19, 20], and PEG10 [21], which in turn further blocks the AR signaling pathways and promotes the emergence of the NE phenotype. Epigenetic changes have been suggested to play a key role in NEPC emergence and/or maintenance [14, 22, 23], although these changes have been poorly studied. Aberrations in the activity of master epigenetic modifiers, including REST [19, 24, 25], EZH2 [14, 18], and HP1α [26], have been shown to be important in the emergence and lineage plasticity of NEPC conferred by appropriate epigenetic modulations, and may facilitate ADT resistance and NEPC transformation. In prostate cancer, lysine-specific demethylase 1 (LSD1) has been shown to promote AR-dependent transcription via its demethylase activity [27,28,29,30]. Recently, LSD1 has been reported to collaborate with ZNF217 to activate a lethal gene network in a noncanonical, demethylase-independent manner in CRPC [31]. LSD1 inhibitors have antitumor effects in vitro and in vivo in small cell lung cancer, and SOX2 overexpression confers sensitivity to LSD1 inhibitors in cancer cells [32, 33].
Previous studies observed that decreased expression of the dominant adenocarcinoma specific genes was accompanied by the onset of SOX2 expression began weeks before the emergence of the neuronal expression signature and persisted through terminal NEPC [21]. This finding highlights the potential crucial roles of SOX2 in driving NEPC transdifferentiation. SOX2 is a SOX family member that belongs to the SOXB1 group and is typically considered to be a key regulator in maintaining the pluripotency and self-renewal properties of embryonic stem cells [34,35,36,37,38]. SOX2 expression has been shown to be lower in neoplastic prostatic epithelium than in normal prostate epithelium, and SOX2 and the NED marker chromogranin-A are primarily coexpressed in both prostate cancer and lymph node metastases [39]. Recent studies have indicated that SOX2 levels are significantly higher in CRPC tumors with neuroendocrine-like histology than in adenocarcinomas [40, 41] and that SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer [40]. However, the role of SOX2 during the transformation from adenocarcinoma to NEPC is still unclear.
In this study, by analyzing the RNA sequencing data of a patient-derived xenograft model and human CRPC data, we showed that SOX2 may serve as a potential repressor of the AdPC phenotype. Furthermore, we characterized the SOX2 response program in vitro through an integrative expression profiling analysis and confirmed that SOX2 may drive the increased incidence of NEPC by inhibiting AdPC master regulators (MRs) through the epigenetic regulator-LSD1. In addition to the reverse suppressive effect on the AdPC phenotype, we also confirmed that SOX2 alone plays a positive but relatively weak role in driving NE differentiation.
Materials and methods
Cell culture and transfection
LNCaP and CWR22RV1 cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and maintained in RPMI 1640 medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin at 37 °C under a humidified atmosphere containing 5% CO2. For gene function studies, exogenous SOX2 was transfected with a lentiviral vector (GV358-EGFP-Puro, GeneChem, Shanghai, China) after the parental cells reached 40–60% confluence, and the transfected cells were selected with puromycin (2 mg/ml) for 5 days. Cells with REST expression knockdown were established by transfection with REST-shRNA lentiviral particles (H5739-mCherry-Amp, Obi Technology, Shanghai, China) following the manufacturer’s protocol. Transfection efficiency was monitored using the corresponding fluorescent protein expression vector. The SOX2 and REST shRNA target sequences were: 5′-AGAAGGATAAGTACACGCT CTCGAG AGCGTGTACTTATCCTTCTTC-3′ and 5′-GCTGGCAAATGTGGCCTTATTCAAGAGATAAGGCCACATTTGCCAGC-3′ respectively.
Quantitative real-time PCR (qRT-PCR)
The mRNA levels of the genes assessed in this study were determined by qRT-PCR assays. Total RNA was extracted from cultured cells by using TRIZOL reagent following the manufacturer’s protocol, and cDNA was prepared by using a first-strand cDNA synthesis kit (TaKaRa Bio, Japan). qRT-PCR was performed in a LightCycler 480 Thermocycler (Roche Applied Science, Mannheim, Germany) using a SYBR Green qPCR kit (TaKaRa Bio, Japan), and the assays were carried out using three independent replicates. The primer sequences used in this study are listed in Table S7.
Antibodies and immunoblot analysis
For immunoblot analysis, total cellular protein was extracted with a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime, China), and equal amounts (20 µg) of lysate were loaded into each lane, separated by 10% SDS-polyacrylamide gel electrophoresis, and then transferred to a polyvinylidene fluoride membranes by electroblotting. After blocking, the protein-transferred membranes were incubated overnight at 4 °C with antibodies against SOX2, FOXA1, NKX3-1, LSD1, H3K4me2, H3K4me1, H3K9me1, SYP, or ASCL1 (all 1:1000, Abcam, Germany); PSA (1:1000, Proteintech, China); or β-actin (1:5000, Beyotime, China). β-Actin served as the loading control, and the bands were detected by chemiluminescence using ECL reagents.
RNA sequencing and analysis
Total RNA was extracted from LNCaP cells expressing SOX2 or from lentivirus-empty vector LNCaP cells with TRIZOL (TaKaRa Bio, Japan), which was followed by an additional purification step. RNA quality was determined by Qubit, and the RNA integrity was verified with an Agilent 2100 Bioanalyzer. After constructing capture libraries and ligating barcoded sequencing adapters, transcriptome sequence data processing and analysis were performed using pipelines at the Biomarker Technologies Company with an Illumina HiSeq platform to generate 40 Gb of clean data. Reads (FASTQ files) were mapped to a human genome reference sequence (hg19). Expression levels (FPKM) were estimated by counting all nucleotides mapped to each gene and normalized to the total number of mapped nucleotides (per million) and the gene length (per kb). Differential expression analysis was performed using DESeq. The genes that showed a fold change of >1.5 and a Benjamini–Hochberg (BH, FDR) correction of <0.05 were considered to be differentially expressed.
Chromatin immunoprecipitation sequencing (ChIP-Seq)
LNCaP cells with or without exogenous SOX2 transfection were collected to perform a ChIP analysis by using a SimpleChIP® Plus Enzymatic Chromatin IP kit (Cell Signaling Technology, MA, USA). Approximately 1 × 107 cells were fixed with formaldehyde to cross-link histone and nonhistone proteins to DNA. Chromatin was digested with micrococcal nuclease into 150-900-bp DNA/protein fragments. The chromatin was then subjected to immunoprecipitation using antibodies specific for the SOX2 protein (Cell Signaling Technology, MA, USA) or normal rabbit IgG, and the complexes were coprecipitated and captured with protein G agarose or protein G magnetic beads. The precipitated immunocomplex crosslinking was reversed with proteinase K, and DNA was purified using spin columns. The purified DNA was sequenced according to the principle using an Illumina HiSeq2500 instrument at the Biomarker Technologies Company after undergoing end repair, adapter ligation, and amplification. Clean reads were mapped to a human genome reference sequence (hg19) using Bowtie2 software, which allowed for two mismatches. Sequence reads that mapped to multiple sites in the human genome were removed. Peak calling was performed by using the MACS2 program, and a p value of <0.001 was used to select the SOX2 peaks using the IgG-ChIP assay data as a control.
Activity of the LSD1
LSD1 activity in LNCaP cells and CWR22RV1 cells was assessed using a KDM1/LSD1 Activity Quantification kit (Abcam, Germany) following the manufacturer’s instructions. The nuclear extracts were generated by using a Nuclear and Cytoplasmic Protein Extraction kit (Beyotime, China). Equal amounts (10 µg) of nuclear extracts were incubated with dimethylated histone H3K4 substrate, which was stably coated onto the strip wells. Active LSD1 binds to this substrate and removes methyl groups from this substrate. The LSD1-demethylated products were recognized with a capture antibody. After incubation with the detection antibody, the ratio or amount of demethylated products, which is proportional to the LSD1 activity, was then calorimetrically measured by reading the absorbance with a colorimetric microplate reader at a wavelength of 450 nm.
Immunofluorescence (IF)
For immunofluorescence, cells were cultured in six-well plates until reaching 60–70% confluence and then were fixed with 4% paraformaldehyde for 15 min at room temperature. After washing in PBS and blocking with goat serum, the cells were stained with an anti-LSD1 antibody (1:50, Abcam, Germany). Subsequently, the primary antibody was visualized with Alexa Fluor® 594-conjugated goat anti-rabbit IgG (1:2000, Proteintech, China). Cell nuclei were stained with DAPI, and fluorescence images were acquired on a High Concept Cell Imagine Instrument(ImageXpress® Pico, Molecular Devices).
Cell proliferation assays
Cell proliferation was analyzed using an EdU assay kits (Ruibo, Guangzhou, China) following the manufacturer’s instructions. Cell proliferation analysis was conducted by plating 2 × 105 LNCaP-SOX2 or LNCaP cells per well in six-well plates in androgen starvation medium with or without dihydrotestosterone DHT. After 3 days, the culture medium was replaced with medium containing EdU, and the cells were cultured for 4 h at 37 °C in a CO2 incubator before performing the cell proliferation assay was performed. The EdU-labeled cells were then fixed with 4% paraformaldehyde and after Apollo and nuclear staining, images of the proliferating cells undergoing DNA replication were captured using a High Concept Cell Imagine Instrument (ImageXpress® Pico, Molecular Devices).
Statistics
The data are presented as the mean ± S.D. All statistical analyses were performed using SPSS 24.0. P-values were calculated using Student’s t-test to determine differences between two groups, and significance at p < 0.05, p < 0.01. p < 0.001 and p < 0.0001 was denoted as *, **, ***, and ****, respectively.
Results
SOX2 is predicted to be the candidate regulator of AdPC specific genes during the NEPC progression
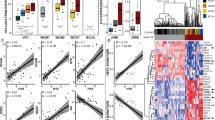
Transcription factors (TFs) function as MRs of lineage development in multiple tissues. To identify the potent MR of the transformation from AdPC to NEPC, we examined the RNA sequencing data from the Beltran et al. dataset [11] and the Akamatsu et al. dataset [21, 42] for transcriptional gene set analysis of NEPC versus AdPC using Enrichr CHEA2016 (http://amp.pharm.mssm.edu/Enrichr/) [43]. Strikingly, SOX2 was predicted to be the top driver TF for the downregulated genes (Tables S1 and S2) associated with the AdPC phenotype (Figs. 1a, b), ranking even higher than the well-established AdPC MRs, such as AR, FOXA1, and NKX3-1 [44,45,46,47]. In the LTL331 xenograft model, the upregulation of SOX2 mRNA expression was also negatively associated with the loss or downregulation of the AdPC specific genes AR, FOXA1, NKX3-1, and KLK3 during the transition from AdPC to NEPC (Fig. 1e).
Identification of driver genes for the progression of NEPC. a, b TF analysis of genes with downregulated expression in the Beltran et al. [11] dataset and the Akamatsu et al. [21] dataset using Enrichr, depicted as combined scores (z-score × −log(p value)), respectively; c, d TF analysis of the genes with upregulated expression in the Beltran et al. [11] dataset and the Akamatsu et al. 21] dataset using Enrichr, respectively; e Negative correlation between SOX2 and AdPC MRs in the NEPC specimens from the Akamatsu et al. [21] dataset; f The best set of TFs predicted by the Mogrify analysis for the conversion of prostate epithelial cells to neural stem cells
Interestingly, SOX2 was also predicted to be a driver TFs of the upregulated genes (Tables S3 and S4) associated with the NEPC phenotype by Enrichr-CHEA 2016, although with much weaker significance than was observed for the other TFs (Fig. 1c, d). SOX2 and BRN2/POU3F2 have been reported to be master TFs for inducing neuronal stem cell transition [35, 48, 49], and the Mogrify algorithm (http://www.mogrify.net) predicted that SOX2, together with BRN2/POU3F2, was the top ranking MR that could induce the lineage transition from prostate epithelial cells to neural stem cells (Fig. 1f), a process presumably similar to that of the ADT-induced transition from AdPC to NEPC. Collectively, these findings indicate that SOX2 plays a potent repressor role in the loss of the AdPC phenotype, and had a positive but relatively weak effect on the terminal establishment of the NEPC phenotype during the progression of NEPC.
Overexpression of SOX2 suppresses the expression of AdPC specific genes
We established stable SOX2 overexpression subline of LNCaP and CWR22RV1 cells (termed LNCaP-SOX2 and CWR22RV1-SOX2) using a lentiviral vector system (Fig. 2a, b) to investigate the effect of SOX2 on the expression of AdPC specific genes. We observed a rapid morphological alteration in the LNCaP-SOX2 cells by 96 h after SOX2 lentiviral transfection (Fig. S1). The qRT-PCR results verified that elevated SOX2 expression caused a 44.96% decrease in AR mRNA expression in cells treated with DHT but had no detectable effects on cells grown in androgen-depleted medium (Fig. 2c). The qRT-PCR results also showed that the expression of other AdPC specific genes, including NKX3-1, PSA, and ADAM7 (Fig. 2d–f), were significantly lower in LNCaP-SOX2 cells than in control cells. Consistently, the decreasing trends in the expression levels of NKX3-1 and PSA in LNCaP cells, and AR and FOXA1 in CWR22RV1 cells after SOX2 transfection were also confirmed at the protein level by western blot analysis (Fig. 2g). In addition, a SOX2 recovery experiment in which SOX2 was knocked down in LNCaP-SOX2 cells with a specific short hairpin RNA (shRNA) resulted in the recovery of AR, NKX3-1, and PSA expression at the protein level (Fig. 2h). Taken together, these results confirmed the suppressive effect of SOX2 on the expression of AdPC specific genes.
SOX2 overexpression suppresses the expression of AdPC specific genes in androgen-responsive cell lines. a, b The mRNA and protein levels of SOX2 in LNCaP and CWR22RV1 cells were detected by qRT-PCR and western blotting; c–f Cells in CSS medium were treated with or without 10 nM DHT. The relative gene expression of AR, NKX3-1, PSA, and ADAM7 in LNCaP cells was analyzed by qRT-PCR over 48 h. Cells in FBS medium were used as the positive control; g The impact of SOX2 on the expression of AR, NKX3-1, FOXA1, and PSA at the protein levels in LNCaP and CWR22RV1 cells was analyzed by western blotting; h LNCaP-SOX2 cells were transfected with a specific shRNA targeting SOX2 or scrambled control shRNA. Western blot analysis showed the protein levels of SOX2, AR, NKX3-1, and PSA. Abbreviation: Parental, lentivirus-free LNCaP cells; DHT, dihydrotestosterone
SOX2 is involved in the epigenetic regulation of AdPC specific genes
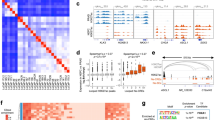
To characterize the SOX2 response program at the global transcriptome level and elucidate the molecular mechanism of the SOX2-driven suppression of the AdPC signature, we performed RNA sequencing of the LNCaP-SOX2 cells and control cells. The RNA sequencing data revealed that 1295 annotated genes were differentially expressed, including 889 upregulated and 406 downregulated genes in the LNCaP-SOX2 cells compared with those in the control cells (DEseq, fold change ≥ 1.5 and FDR ≤ 0.05). As shown in Fig. 3a, the LNCaP-SOX2 cells exhibited low expression of AdPC specific genes (e.g., AR, NKX3-1, KLK3, FKBP5, TMEFF2, and ADAM7) and high expression of only a few neuroendocrine-associated genes (e.g., AURKA, PEG10, CENPE, SYT4, and ENO2) and pluripotency genes (e.g., CD24 and KLF4). The reliability of the RNA sequencing data was confirmed by qRT-PCR analysis of eight downregulated genes (NKX3-1, NDRG1, NTNG1, TMEFF2, MAF, ADAM7, MCCC2, and CAMK2N1) and three upregulated genes (KLF4, MK167, and FOXM1), which produced consistent results (Fig. 3b). Pathway and process enrichment analysis of the 406 genes with decreased expression (Table S5) using Metascape (http://metascape.org/gp/index.html) [50], which is a collection of pathways from various databases, revealed that the pathways of “activated PKN1 stimulates transcription of AR regulated genes KLK2 and KLK3” (R-HSA-5625886), “PRC2 methylates histones and DNA” (R-HSA-212300), “epigenetic regulation of gene expression” (R-HSA-212165), “RMTs methylate histone arginines” (R-HSA-3214858), and “regulation of epithelial to mesenchymal transition” (GO:0010717) were significantly enriched (p < 0.01) (Fig. 3c). Enrichr transcriptional gene set analysis showed that AR and FOXA1 were the master TFs of the 406 downregulated genes in LNCaP-SOX2 cells (Fig. 3d).
The molecular features of SOX2 in LNCaP cells. a Heat map showing the differentially expressed genes between the LNCaP-SOX2 cells and control cells; b Confirmation of the RNA sequencing results by qRT-PCR; c Enrichment analysis of the genes with lower expression in LNCaP-SOX2 cells than in control cells using Metascape; d TF analysis of the genes with lower expression in LNCaP-SOX2 cells than in control cells using Enrichr; e DNA binding sequence distribution of SOX2 in LNCaP-SOX2 cells, as measured by ChIP-Seq
To determine whether SOX2 suppresses the expression of AdPC specific genes through a direct binding mechanism, we further performed ChIP-Seq with a monoclonal antibody specific for SOX2 in LNCaP-SOX2 cells, which resulted in the identification of 2115 significantly enriched SOX2 binding regions in the human genome (MACS, p < 0.001). Among these regions, only 2.46% mapped to within 3 kilobases (kb) of gene transcriptional start sites, and none mapped to the transcriptional regulatory region of the genes in our defined AdPC specific gene sets (Fig. 3e). Taken together, the results of the RNA sequencing and ChIP-Seq data suggested that SOX2 is more likely to suppress the AdPC specific genes through epigenetic modifications rather than a direct binding mechanism.
LSD1 is a crucial target of SOX2 and may contribute to the global hypomethylation of AdPC specific genes
Our observation that increased SOX2 expression decreased the expression of the gene set associated with the epigenetic regulation of gene expression and that the majority of the SOX2 enriched regions mapped to distal intergenic sites in LNCaP-SOX2 cells prompted us to investigate the functional relationship between SOX2 and the epigenetic changes in androgen-responsive prostate cancer cell lines. Strikingly, we observed that SOX2 overexpression induced dramatic global hypomethylation of both histone H3 at lysine 4 (H3K4) and histone H3 at lysine 9 (H3K9) in both LNCaP and CWR22RV1 cells compared with that observed in control cells (Fig. 4a). Given that LSD1 is a well-established lysine-specific demethylase that plays an important role in histone modification dynamics by removing the di- and mono-methylation marks from methylated H3K4 (H3K4me1/me2) to suppress gene expression and from H3K9 (H3K9me1/me2) to promote gene expression [27, 51], we next examined the impact of SOX2 on LSD1 expression. We observed that SOX2 overexpression significantly enhanced the total protein levels, nuclear protein levels, and enzymatic activity of LSD1 in LNCaP cells and increased the LSD1 protein levels in CWR22RV1 cells (Fig. 4a–c). Interestingly, SOX2 overexpression did not significantly change the level of LSD1 mRNA expression in LNCaP-SOX2 cells (Fig. S2).
SOX2 reduces the expression of prostate AdPC specific genes via LSD1. a The indicated cells were cultured in CSS medium or in FBS medium with or without 10 nM DHT for 48 h. Histone methylation and the total LSD1 were analyzed by western blotting; b LNCaP cells were cultured in CSS medium with or without 10 nM DHT for 48 h, and were then fixed and immunostained with an anti-LSD1 antibody. Nuclear LSD1 was examined by immunofluorescence (IF); c LNCaP cells were cultured in CSS medium or in FBS medium with or without 10 nM DHT for 48 h. LSD1 enzymatic activity was measured using a KDM1A/LSD1 Activity Quantification kit (colorimetric); d The indicated cells were cultured in FBS medium with or without 3 mM pargyline for 48 h. The enzymatic activity of LSD1 was measured using a KDM1A/LSD1 Activity Quantification kit and the impact of an LSD1 inhibitor on the expression of AdPC specific genes and the methylation levels of H3K9 and H3K4 was analyzed by western blotting
To assess whether the decrease in AdPC specific gene expression was mediated by SOX2 through an LSD1-dependent mechanism, we evaluated the impact of a chemical inhibitor of LSD1, pargyline, on the expression of AdPC specific genes. Interestingly, we observed that the LSD1 inhibitor treatment induced further decreases in the expression of the AdPC specific TFs AR and NKX3-1 and the AR target gene PSA in LNCaP cells, and of AR and FOXA1 in CWR22RV1 cells, as assessed by western blotting (Fig. 4d). The results of the western blot analysis showed that this effect of was probably attributable to the reversion of H3K4 and H3K9 methylation driven by the LSD1 inhibitor (Fig. 4d).
Furthermore, in LNCaP cells, we detected a correlation between the expression of SOX2 and EZH2, the latter of which has recently been identified as the master epigenetic modifier in NEPC samples and is implicated in NEPC progression [11, 18, 52]. However, no detectable effects of SOX2 on EZH2 expression or of the EZH2 inhibitor-GSK126 on the expression of AdPC specific genes were observed in LNCaP-SOX2 cells and control cells (Fig. S3).
SOX2 alone weakly induces the NE-like phenotype
SOX2 is sufficient to confer a cell lineage plasticity status [40]. Consistently, our RNA sequencing data showed the upregulated expression of some NEPC driver genes (such as AURKA, PEG10, and EZH2) and a few NE markers (such as ENO2, SYP, and SYT4), but not global acquisition of the NEPC phenotype in LNCaP-SOX2 cells (Fig. 3a). As shown in Fig. 5a, b, and d, SOX2 overexpression moderately increased the expression of the canonical NE markers ENO2 and SYP at the mRNA level and that of SYP and ASCL1 at the protein level in LNCaP cells, and of ENO2 and SYP at the protein level in CWR22RV1 cells. Furthermore, when REST, the master repressor of NE genes was knocked down in LNCaP-SOX2 cells, the expression of SYP, SCG3, and ASCL1 increased dramatically (Fig. 5c, d).
SOX2 induces neuroendocrine differentiation. a, b LNCaP cells were cultured in FBS medium for 48 h, and the relative gene expression of ENO2 and SYP was measured by qRT-PCR; c LNCaP cells were transfected with a short hairpin RNA (shRNA) targeting REST or with a vehicle control. qRT-PCR was performed to assess the mRNA expression of SCG3; d Western blot analysis was performed to measure the expression of NE markers in LNCaP and CWR22RV1 cells; e Enrichment analysis of the genes exhibiting higher expression in LNCaP-SOX2 cells than in control cells using Metascape; f Enrichr analysis of master TFs for the gene set upregulated in LNCaP-SOX2 cells compared with those observed in control cells; g LNCaP cells in CSS medium were treated with 10 nM DHT for 48 h, and cell proliferation was measured using an EdU assay kit
Clinically, NEPC is also characterized by an NE-like phenotype and a highly aggressive and proliferative phenotype [6, 7]. SOX2 overexpression resulted in the upregulation of gene panels involving “cell cycle, mitotic” (R-HSA-69278), “DNA replication” (GO:0006260) and “FOXM1 pathway” (M176) from our RNA sequencing data (Fig. 5f). Furthermore, our results showed that FOXM1, which is a pro-proliferative TF that promotes cell cycle progression at G1-S, and G2-M transitions [53], exhibited the highest combined score for the increased genes in the Beltran et al. [11] dataset (Fig. 1g), and our RNA sequencing data (Fig. 5e) provided further support for its essential role in the aggressive proliferation of NEPC. However, no measurable differences in cell proliferation between LNCaP-SOX2 cells and control cells were detected (Fig. 5g). Considering that SOX proteins function mainly through forming complexes with partner proteins and binding to DNA sequences [54], we considered that an additional aberrations, in addition to SOX2 alteration, may be required for further AdPC transdifferentiation from the SOX2-induced plasticity status of prostate cancer to the canonical NEPC status.
Discussion
Critical issues associated with NEPC include identifying molecular drivers of the transition to this highly aggressive variant of CRPC from AdPC and the development of novel therapeutic options for treating NEPC patients [5, 6, 55].
Transcriptional gene set analysis of RNA sequencing data from the Beltran et al. [11] dataset and the Akamatsu et al. [21] dataset indicated that SOX2 is a potential repressor that drives the decreased expression of adenocarcinoma specific genes during the progression of NEPC. This repressor role of SOX2 was subsequently confirmed by global transcriptome profiling of our LNCaP-SOX2 and CWR22RV1-SOX2 models in vitro. Notably, FOXA1, NKX3-1, and AR, the three MRs of the AdPC luminal phenotype and inhibitors of the NEPC phenotype [44,45,46,47, 56], are the targets of SOX2 repression in the SW620 cell line. In future experiments, it will be interesting to determine whether SOX2 can directly repress AdPC gene expression in NEPC in vivo.
SOX2 has been shown to promote castration-resistant disease and lineage plasticity in TP53- and RB1-deficient prostate cancer [14, 16, 40], which may contribute to the biological heterogeneity of prostate cancer and correlate with prostate cancer aggressiveness through lineage switching. In this study, we confirmed that increased SOX2 expression can alter the epithelial morphology of LNCaP cells and induce the expression of pluripotency genes, indicating that increased SOX2 expression destabilizes the state of epithelial differentiation in LNCaP cells. Furthermore, because the overexpression of SOX2 could sharply inhibit the expression of adenocarcinoma-specific MRs (FOXA1, NKX3-1, and AR) and some AR target genes in the androgen-responsive prostate cancer cell lines; thus, we concluded that SOX2 induction may remove the impetus for differentiation toward the CRPC lineage and thereby facilitate differentiation toward the NEPC lineage.
Our data also revealed that increased SOX2 expression induced significant global hypomethylation of H3K4me1/2 and H3K9me1/2 in both LNCaP and CWR22RV1 cells, an epigenetic changes that are typically driven by the demethylase LSD1. We subsequently demonstrated that SOX2 could potently enhance the expression and enzymatic activity of LSD1 in prostate cells in vitro. Human NEPC exhibits increased expression of stem cell reprogramming factors and epigenetic regulators [14, 57], and epigenetic modifiers may play a role in the induction and/or maintenance of the treatment-resistant status of NEPC [11]. The results presented in this study support the hypothesis that increased SOX2 expression in prostate cancer cells alters the landscape of histone methylation, which creates a stem cell-like epigenetic environment that permits lineage plasticity. In addition, we confirmed that opposing LSD1-mediated demethylation with an LSD1 inhibitor dramatically inhibited of H3K9 and H3K4 methylation in both LNCaP cells and CWR22RV1 cells. The observed decrease in the expression of the AdPC specific genes in this study is likely attributable to the reversion of histone methylation levels driven by the LSD1 inhibitor. However, the extent of LSD1 demethylation and the identity of the dominant substrates underlying this process are still largely unknown and warrant further study.
Recent studies have shown that SOX2 levels are significantly higher in CRPC tumors with NE-like histology than in adenocarcinomas [11, 21], indicating a potential role for SOX2 in the initiation of the NE phenotype. In our SOX2 overexpression cell line model, we also confirmed the weak impact of SOX2 alone on the induction of NE markers, consistent with previous reports [39, 40] and indicating that additional aberrations, such as TP53 and RB1 genetic loss, BRN2 expression, or REST silencing, are required to promote the induction of the NE phenotype. Concurrent TP53 and RB1 alterations are present in 74% of metastatic CRPCs with neuroendocrine-like histology [40]. Furthermore, conditional TP53 and RB1 deletion in mice is sufficient to generate neuroendocrine-like prostate cancer [58]. The wild-type of TP53 and RB1 in LNCaP cells and heterozygous TP53 mutation in CWR22RV1 cells may be the primary block for the progression of NEPC. Notably, an unique hybrid luminal-neuroendocrine tumor that has a neuroendocrine phenotype but still exhibits robust expression of AdPC specific genes and does not progress to terminal NEPC is SOX2-negative [59]. Interestingly, in this study, AR gene expression remained robust despite the decreased expression of many AR signaling genes. We speculated that these results were likely observed because the SOX2 overexpression model represents the transient state between CRPC and NEPC, which has some morphological features of neuroendocrine carcinoma but mixed luminal and basal epithelial characteristics.
In summary, we identified SOX2 as a potent regulator of NEPC that markedly suppresses the expression of AdPC specific genes and marginally induced the expression of some NE markers. Furthermore, this effect was mechanistically the result of the epigenetic modulation driven by LSD1 activation, as demonstrated by the results of our in vitro experiments. Future studies using clinical NEPC samples may aid in establishing LSD1 as a novel target for the treatment of NEPC patients with high SOX2 expression.
References
Toivanen R, Shen MM. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development. 2017;144:1382–98.
Shiota M, Yokomizo A, Naito S. Increased androgen receptor transcription: a cause of castration-resistant prostate cancer and a possible therapeutic target. J Mol Endocrinol. 2011;47:R25–41.
Bishop JL, Davies A, Ketola K, Zoubeidi A. Regulation of tumor cell plasticity by the androgen receptor in prostate cancer. Endocr Relat Cancer. 2015;22:R165–82.
Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45.
Yuan T-C, Veeramani S, Lin M-F. Neuroendocrine-like prostate cancer cells: neuroendocrine transdifferentiation of prostate adenocarcinoma cells. Endocr Relat Cancer. 2007;14:531–47.
Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–95.
Terry S, Beltran H. The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol. 2014;4:60.
Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29:536–47.
Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78.
Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, et al. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res. 2014;20:890–903.
Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305.
Aparicio AM, Shen L, Tapia EL, Lu JF, Chen HC, Zhang J, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res. 2016;22:1520–30.
Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28.
Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83.
Li Y, Donmez N, Sahinalp C, Xie N, Wang Y, Xue H, et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2017;71:68–78.
Yu J, Kregel S, Kiriluk KJ, Rosen AM, Cai Y, Reyes EE, et al. Sox2 is an androgen receptor-repressed gene that promotes castration-resistant prostate cancer. PLoS ONE. 2013;8:e53701.
Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71.
Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30:563–77.
Zhang X, Coleman IM, Brown LG, True LD, Kollath L, Lucas JM, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21:4698–708.
Lee AR, Gan Y, Tang Y, Dong X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine. 2018;35:167–77.
Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015;12:922–36.
Crea F, Venalainen E, Ci X, Cheng H, Pikor L, Parolia A, et al. The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer. Epigenomics. 2016;8:721–31.
Clermont PL, Lin D, Crea F, Wu R, Xue H, Wang Y, et al. Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin Epigenetics. 2015;7:40.
Flores-Morales A, Bergmann TB, Lavallee C, Batth TS, Lin D, Lerdrup M, et al. Proteogenomic characterization of patient-derived xenografts highlights the role of REST in neuroendocrine differentiation of castration-resistant prostate cancer. Clin Cancer Res. 2019;25:595–608.
Chen R, Li Y, Buttyan R, Dong X. Implications of PI3K/AKT inhibition on REST protein stability and neuroendocrine phenotype acquisition in prostate cancer cells. Oncotarget. 2017;8:84863–76.
Ci X, Hao J, Dong X, Choi SY, Xue H, Wu R, et al. Heterochromatin protein 1alpha mediates development and aggressiveness of neuroendocrine prostate cancer. Cancer Res. 2018;78:2691–704.
Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 2014;9:1618–27.
Liang Y, Ahmed M, Guo H, Soares F, Hua JT, Gao S, et al. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res. 2017;77:5479–90.
Ketscher A, Jilg CA, Willmann D, Hummel B, Imhof A, Russeler V, et al. LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogenesis. 2014;3:e120.
Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9.
Sehrawat A, Gao L, Wang Y, Bankhead A 3rd, McWeeney SK, King CJ, et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci USA. 2018;115:E4179–e88.
Zhang X, Lu F, Wang J, Yin F, Xu Z, Qi D, et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013;5:445–57.
Takagi S, Ishikawa Y, Mizutani A, Iwasaki S, Matsumoto S, Kamada Y, et al. LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res. 2017;77:4652–62.
Jia X, Li X, Xu Y, Zhang S, Mou W, Liu Y, et al. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol. 2011;3:230–8.
Zhang S. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J Stem Cells. 2014;6:305–11.
Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med. 2014;3:19.
Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007;20:474–81.
Johansson H, Simonsson S. Core transcription factors, Oct4, Sox2 and Nanog, individually form complexes with nucleophosmin (Npm1) to control embryonic stem (ES) cell fate determination. Aging. 2010;2:815–22.
Russo MV, Esposito S, Tupone MG, Manzoli L, Airoldi I, Pompa P, et al. SOX2 boosts major tumor progression genes in prostate cancer and is a functional biomarker of lymph node metastasis. Oncotarget. 2016;7:12372–85.
Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–8.
Yu X, Cates JM, Morrissey C, You C, Grabowska MM, Zhang J, et al. SOX2 expression in the developing, adult, as well as, diseased prostate. Prostate Cancer Prostatic Dis. 2014;17:301–9.
Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2013;74:1272–83.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7.
Gao N, Ishii K, Mirosevich J, Kuwajima S, Oppenheimer SR, Roberts RL, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–43.
Kim J, Jin H, Zhao JC, Yang YA, Li Y, Yang X, et al. FOXA1 inhibits prostate cancer neuroendocrine differentiation. Oncogene. 2017;36:4072–80.
Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–77.
Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32:399–414.
Urban S, Kobi D, Ennen M, Langer D, Le Gras S, Ye T, et al. A Brn2-Zic1 axis specifies the neuronal fate of retinoic-acid-treated embryonic stem cells. J Cell Sci. 2015;128:2303–18.
Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–3.
Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18:723–35.
Maiques-Diaz A, Somervaille TC. LSD1: biologic roles and therapeutic targeting. Epigenomics. 2016;8:1103–16.
Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nature reviews Urology. 2018;15:271–86.
Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–33.
Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–44.
Galletti G, Leach BI, Lam L, Tagawa ST. Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treat Rev. 2017;57:16–27.
Talos F, Mitrofanova A, Bergren SK, Califano A, Shen MM. A computational systems approach identifies synergistic specification genes that facilitate lineage conversion to prostate tissue. Nat Commun. 2017;8:14662.
Nelson EC, Cambio AJ, Yang JC, Ok JH, Lara PN Jr., Evans CP. Clinical implications of neuroendocrine differentiation in prostate cancer. Prostate Cancer Prostatic Dis. 2007;10:6–14.
Zhou Z, Flesken-Nikitin A, Corney DC, Wang W, Goodrich DW, Roy-Burman P, et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 2006;66:7889–98.
Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol. 2012;227:53–61.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81372775).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, H., Wang, L., Li, Z. et al. SOX2 has dual functions as a regulator in the progression of neuroendocrine prostate cancer. Lab Invest 100, 570–582 (2020). https://doi.org/10.1038/s41374-019-0343-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0343-5
This article is cited by
-
Alternative splicing in prostate cancer progression and therapeutic resistance
Oncogene (2024)
-
Epigenetic mechanisms underlying subtype heterogeneity and tumor recurrence in prostate cancer
Nature Communications (2023)
-
Advances in neuroendocrine prostate cancer research: From model construction to molecular network analyses
Laboratory Investigation (2022)
-
SOX2 mediates metabolic reprogramming of prostate cancer cells
Oncogene (2022)
-
Molecular mechanisms of neuroendocrine differentiation in prostate cancer progression
Journal of Cancer Research and Clinical Oncology (2022)