Abstract
To date, more than 170 chemical modifications have been characterized in RNA, providing a new layer of gene expression regulation termed the ‘epitranscriptome’. RNA modification detection methods and tools advance the functional studies of the epitranscriptome. According to the detection throughput and principles, existing RNA modification detection technologies can be categorized into four classes, including quantification methods, locus-specific detection methods, next-generation sequencing-based detection technologies and nanopore direct RNA sequencing-based technologies. In this review, we summarize the current knowledge about these RNA modification detection technologies and discuss the challenges for the existing detection tools, providing information for a comprehensive understanding of the epitranscriptome.
Similar content being viewed by others
Introduction
RNA is a single-stranded molecule consisting of four nucleotides: adenosine (A), guanosine (G), cytidine (C) and uridine (U). RNA, both an important and conserved macromolecule, not only participates in the flow of genetic information but also regulates gene expression. Beyond sequence information, chemical modifications add to the complexity of RNA, emerging as a new layer of gene expression regulation. Since the first chemical modification was characterized 60 years ago, more than 170 RNA modifications have been characterized1. Most of the RNA modifications have been identified in abundant non-coding RNAs, including ribosomal RNA (rRNA), transfer RNA (tRNA), and small nuclear RNA (snRNA).
The development of detection technologies advances the investigation of the functional roles of RNA modifications2,3,4,5,6,7. To date, more than ten chemical modifications have been mapped in a transcriptome-wide manner, including N6-methyladenosine (m6A), N6, 2′-O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), inosine (I), pseudouridine (Ψ), N1-methyladenosine (m1A), 2ʹ-O-methylation (Nm), N4-acetylcytidine (ac4C), N7-methylguanosine (m7G) and dihydrouridine (D). Different chemical modifications play distinct regulatory roles in RNA metabolism and function. For instance, m6A, the most abundant internal messenger RNA (mRNA) modification, influences RNA metabolism in multiple ways, including stability, splicing, translation, localization and RNA secondary structure8,9,10,11. m5C in mRNA influences mRNA export, RNA stability, and translation, and m5C in tRNA is essential for maintaining structural stability and translational fidelity12,13,14,15,16,17,18,19. Inosine preferentially exists in double-stranded RNA (dsRNA) regions and affects codon recoding, splice-site choice, microRNA (miRNA) biogenesis and targeting efficiency20,21,22. Ψ is required for proper rRNA folding, tRNA structure stabilization and snRNP (small nuclear ribonucleoprotein) biogenesis23,24,25,26,27,28,29,30,31,32. In addition, introducing Ψ into mRNA increases protein production and alters translation33,34,35. m1A at position 58 in tRNA is conserved and vital for stabilizing tRNA tertiary structure, and m1A in mRNA influences translation36,37,38,39,40,41,42,43,44,45,46,47. Nm is essential for accurate and efficient protein synthesis48,49,50,51,52. Internal m7G increases mRNA translation efficiency and augments miRNA biogenesis53,54. ac4C in mRNA promotes translation, and ac4C in rRNA can affect rRNA biogenesis55,56,57,58,59.
RNA modification detection technologies provide not only resources for a comprehensive understanding of the epitranscriptome but also tools for functional studies. Hence, in this review, we will summarize the current knowledge about these existing RNA modification detection technologies and discuss the challenges for these existing detection tools. The detection technologies are categorized according to the detection throughput and principles into four classes: quantification methods, locus-specific detection methods, next-generation sequencing-based detection technologies and nanopore direct RNA sequencing-based technologies.
RNA modification quantification methods
The identification and quantification of new modified nucleotides requires powerful RNA modification quantification methods. Based on the principle that modified nucleotides possess distinct chemical properties from the originals, several RNA modification quantification methods have been established, including two-dimensional thin-layer chromatography (2D-TLC), dot blot, and liquid chromatography–mass spectrometry (LC–MS). These approaches can be used to quantify the modification abundance in specific RNA species and require highly purified RNA due to the lack of sequence information.
2D-TLC
2D-TLC is a widely used RNA modification detection method according to the distinct mobilities of different nucleotides in the solvent60,61,62. In detail, isolated RNA is first partially digested into oligonucleotides using RNase A, T1, or T2 and then labeled with 32P using T4 polynucleotide kinase (T4 PNK). Finally, 5ʹ-32P-NMP is acquired by nuclease P1 digestion and further separated by 2D-TLC. The nucleotides can be determined by assignment to the standards by comparing their retardation factor (Rf) values (Fig. 1). Quantification of the nucleotides is achieved by measuring the radioactivity of the corresponding spots in the TLC plate63,64,65. This approach is very sensitive and requires only a small amount of RNA (from 50 ng to 200 ng). Consequently, this approach can be applied to both abundant non-coding RNAs (rRNA and tRNA) and less abundant mRNA. In addition, it can also detect RNA modifications in specific tRNA or snRNA sequences, which can be isolated by gel purification or hybridization methods. Furthermore, this approach does not require expensive instruments and thus can be run inexpensively. However, this method also has certain drawbacks, including the requirement of a radioactive reagent, the bias caused by RNase digestion and discrepant 32P labeling efficiency for the modified nucleotides63,66.
Dot blot
Dot blot assay is established and applied to detect and quantify modification levels in RNAs using a specific antibody. In detail, isolated RNAs are stalled to the polyvinylidene fluoride (PVDF) or nitrocellulose membrane directly without electrophoretic size separation. A specific antibody for target modification is incubated with the membrane, followed by secondary antibody hybridization and subsequent signal trapping (Fig. 1). Comparing results from different experimental groups can obtain semiquantitative modification level information. This approach has been widely applied to various RNA species, including non-coding RNAs and mRNAs. Given the detection principle, the sensitivity and accuracy of this approach are highly dependent on the specificity of the antibody. In addition, the amount of starting material, ranging from nanograms to micrograms, also depends on the abundance of the RNA modifications of interest. Since the workflow is straightforward and can be performed inexpensively, this approach has been widely applied for RNA modification detection67,68. However, the lack of absolute quantification and locus information limits a wider application of this approach.
HPLC and LC–MS
High-performance liquid chromatography (HPLC) is an advanced column chromatography technology to separate nucleosides according to their distinct polarities. Prior to HPLC analysis, RNA or oligonucleotides are digested and dephosphorylated to single nucleosides by nuclease P1 and alkaline phosphatase. The UV absorbance and retention time of the nucleosides are recorded by a UV detector, which can be used to identify and measure the abundance of the modified nucleosides (Fig. 1). Compared with 2D-TLC, HPLC analysis is rapid and free of radiolabeling. However, this approach can only be applied to detect highly abundant modifications in abundant RNA species, such as rRNA, tRNA and synthetic RNAs, and requires a large amount of purified RNA (more than 1 µg) due to the detection limit of the UV detector69.
To increase detection sensitivity, HPLC is coupled with mass spectrometry70. Similarly, RNA or oligonucleotides are completely digested to nucleosides and separated by reverse column chromatography. Then, these nucleosides are ionized and further fragmented into specific product ions via mass spectrometry. Integration of retention time, mass-to-charge ratio (m/z) and product ion are capable of determining a certain nucleoside (Fig. 1). In addition, quantification of nucleosides can be achieved through the external standard curve in the same batch60,71. The extremely high sensitivity of triple quadrupole-based mass spectrometry provides this approach a detection limit that can reach the low femtomolar range, and the amount of starting material can be as low as 50 ng, thereby allowing the determination and quantification of these low-abundance modifications in mRNA and low-abundance ncRNA72,73. Hence, LC–MS has been a benchmark for RNA modification detection and quantification. However, the limit of this technique is the requirement of instruments including HPLC and mass spectrometry. In addition, due to the lack of sequence information, when detecting and quantifying RNA modifications in mRNA or other less abundant RNA types, caution must be taken to reduce contamination from abundant and highly modified rRNA and tRNA.
Locus-specific detection methods
The precise position information of a certain RNA modification is important for functional studies. To date, several locus-specific RNA modification detection methods have been developed. These methods can be categorized according to the detection principles into four classes: (1) Primer extension; (2) RNase H-based approaches; (3) electrospray ionization-mass spectrometry (ESI-MS)-based approaches; and (4) semiquantitative PCR- or qPCR-based approaches.
Primer extension
This approach is based on reverse transcription and has been used extensively to detect and localize various RNA modifications, including m1A, Ψ, and m1G. In principle, a 5ʹ-labeled specific RT primer is hybridized with the RNA of interest and extended by reverse transcriptase. Hence, this approach relies on prior knowledge of the modification type and sequence information of the target RNA. Without modifications, reverse transcriptase can reach the 5ʹ end of RNA and generate full-length cDNA. When encountering modified nucleotides, the extension of reverse transcriptase is blocked immediately upstream of the modified site. Then, the RT products are separated using denaturing polyacrylamide gels, and the terminal position of truncated cDNA indicates where the modification occurred (Fig. 2a). This approach is very sensitive and can be applied to various RNA species, including rRNA, tRNA, snRNA and abundant mRNA42,43,74,75,76,77,78,79,80. In addition, owing to the hybridization step, this approach possesses high detection specificity and does not require purified homogenous RNA as the starting material. A major limitation of this method is that it can only detect the modifications or their chemical adducts that are capable of blocking reverse transcription; it is not suitable for RT-silent modifications such as m6A and m5C81,82,83.
a Technical flowchart of primer extension. The truncated cDNA product indicates where the modification occurs. b Technical flowchart of SCARLET, an RNase H-based approach; the modified and unmodified form of the nucleotide of interest can be accurately quantified by TLC; TAP, thermosensitive alkaline phosphatase. c Technical flowchart of RTL-P, a semiquantitative PCR-based approach; Nm impedes RT under low-dNTP conditions and allows readthrough under high-dNTP conditions, and the Nm status can thus be determined by comparing the intensity of the longer PCR product with that of the shorter product. d Technical flowchart of the SELECT, qPCR-based approach; this approach utilizes the feature that m6A hinders the elongation of Bst and reduces the ligation efficiency of SplintR to distinguish modified and unmodified RNA. e Technical flowchart of the qPCR-based Ψ detection approach; this approach utilizes the feature that the Ψ-CMC adducts induce mutation/deletion in cDNA, which alters the melting curves of the qPCR products. f Technical flowchart of the ESI-MS-based approach. Modifications that can be detected by the corresponding approaches are listed below.
RNase H-based approach
The RNase H-based approach is independent of reverse transcription and thus can detect and quantify RNA modifications that do not affect Watson-Crick base pairing. In this method, purified RNA is cleaved into two halves at the 5ʹ end of the nucleotide of interest by RNase H, and cleavage specificity is achieved by annealing with a specific 2′-O-methyl RNA–DNA chimera oligonucleotide. Then, the 3ʹ-half of the RNA is purified, and its 5′ terminus is further labeled with 32P. Furthermore, the oligonucleotides are completely digested into single nucleotides and subsequently resolved by TLC. Owing to the radiolabeling specificity, only the modified and unmodified forms of the nucleotide of interest can be detected and accurately quantified by TLC analysis64,84. This approach is sensitive and powerful for quantifying the stoichiometry of modified nucleotides. However, as the specific 2′-O-methyl RNA–DNA chimera oligonucleotide is base-paired with the target RNA sequence to guide RNase H cleavage, sequence information is needed, and modifications that affect Watson-Crick base pairing cannot be detected and quantified by this method. In addition, to reduce false signals from other RNA sequences, homogenously purified RNA, isolated by the hybridization method and gel purification, is used as the starting material. Therefore, this method requires a large amount of RNA and can be applied to abundant non-coding RNAs, such as rRNA and snRNA64,84.
To quantitatively detect modifications in mRNA/lncRNA, a new method, named SCARLET (site-specific cleavage and radioactive labeling followed by ligation-assisted extraction and thin-layer chromatography), has been developed85. In SCARLET, a 116-nucleotide single-stranded DNA oligonucleotide is splint-ligated to the 32P-labeled RNA fragment of interest and then subjected to RNase T1/A digestion. The splint ligation enables RNA mixtures to be used as the starting material rather than a specific purified RNA, and modifications in mRNA and lncRNA can thus also be quantified. The accurate size of the DNA oligonucleotide is recovered by gel purification. Then, the 32P-labeled nucleotides are released by nuclease P1 digestion and further analyzed by TLC (Fig. 2b). This approach has been applied to quantify m6A and Ψ modification status in mRNA85,86. Since this approach avoids reverse transcription, it can be used as an orthogonal method to validate modification sites identified by transcriptome-wide methods that are dependent on reverse transcription. As with other RNase H-based methods, SCARLET also requires prior knowledge of the sequence information to target a single specific nucleotide and cannot quantify the modification status in a de novo manner. In addition, the requirement of radioactive reagents and complicated procedures limit wider application of this method.
Semiquantitative PCR or qPCR-based approach
Similar to primer extension assays, semiquantitative PCR or qPCR-based approaches are also based on the fact that modified nucleotides impede reverse transcriptase extension. In contrast to primer extension and RNase H-based approaches, qPCR-based approaches are free of radiolabeling and hence are time-saving and easy to perform in the laboratory. To date, several semiquantitative PCR- or qPCR-based approaches have been developed and have successfully detected Nm, Ψ and m6A in diverse RNA species (Fig. 2c–e)87,88,89,90,91,92,93. In addition, owing to the high sensitivity and specificity of PCR and qPCR, these approaches do not require purified RNA as the starting material and can be applied to various RNA species, including less abundant mRNA and lncRNA.
Nm blocks reverse transcriptase extension at a low dNTP concentration and allows readthrough at a high dNTP concentration82,94. Taking advantage of this property, researchers have developed a method referred to as RTL-P (reverse transcription at low dNTP concentrations followed by PCR). In this method, the RNA of interest is first reverse-transcribed at both low and high dNTP concentrations. The truncated and full-length cDNA are amplified by specific primers. The PCR products are analyzed by gel electrophoresis, and the Nm status can be determined by comparing the intensity of the longer PCR product with that of the shorter product (Fig. 2c)87. In addition to altering RT conditions, RT enzymes can also be engineered to facilitate modified nucleotide detection. For instance, an engineered thermostable KlenTaq DNA polymerase variant possesses reverse transcription activity and can discriminate Nm at normal dNTP concentrations. The combination of this engineered DNA polymerase with qPCR has achieved expeditious quantification of Nm88.
In addition to the engineered thermostable KlenTaq DNA polymerase variant, two other DNA polymerases, Tth and Bst, also have reverse transcriptase activity and exhibit distinct capacities to extend when encountering m6A residues versus A residues, which allows locus-specific detection of m6A89,90,91. Moreover, to increase the detection sensitivity of m6A, another method referred to as the single-base elongation- and ligation-based PCR amplification method (SELECT) has been developed92,93. Because m6A can both hinder the elongation activity of DNA polymerases and reduce the nick ligation efficiency of SplintR ligase, in SELECT, cDNA products formed from m6A-containing RNA templates are dramatically reduced, hence significantly increasing detection robustness. Based on qPCR, SELECT is a powerful tool to quantify the m6A fraction in a linear manner (Fig. 2d).
In addition to truncations, the property of induced mutation/deletions by modified nucleotides can also be utilized for locus-specific detection. For instance, Ψ can be selectively labeled by N-cyclohexyl-N′-(2-morpholinoethyl) carbodiimide (CMC), and the Ψ-CMC adducts interfere with Watson-Crick base pairing during reverse transcription. Under optimized RT conditions, Ψ–CMC adducts can be read through and induce mutation/deletion in cDNA. Such mutation/deletions alter the melting curves of the qPCR products, thus enabling the locus-specific detection of Ψ modification (Fig. 2e)95.
ESI-MS-based approach
In contrast to the LC–MS-based nucleoside quantification strategy described above, in this approach, isolated RNA samples are first digested into 5–15 nucleotide fragments by selective endoribonucleases, such as RNase T1, RNase A, and RNase U2. Then, the oligonucleotides are separated by HPLC, and the sequence ladders from the oligonucleotides are further generated through ESI-MS, which can be used for sequence reconstruction and modification identification (Fig. 2f). Hence, this approach can provide both site and stoichiometry information for the modifications of interest70,96,97,98,99. Compared with other locus-specific methods, the ESI-MS-based approach does not rely on prior knowledge of sequence information and thus is able to detect and quantify RNA modification in a de novo manner. Therefore, this approach has been widely applied for modification detection in abundant RNAs, including tRNA, rRNA and snRNA100,101,102,103,104,105,106,107,108,109. Recently, this approach has been used to detect modifications in miRNA and cap modifications in mRNA53,110. The major limitation of this approach is the requirement for highly sensitive ESI-MS. In addition, given the detection principle, the requirement of starting material is large; thus, this method is only suitable for abundant RNA species.
Next-generation sequencing-based detection technologies
With the benefit of advances in next-generation sequencing, an increasing number of RNA modification sequencing technologies have been developed. Such technologies represent powerful tools to map modified nucleotides in a transcriptome-wide manner and promote the elucidation of the regulatory roles of RNA modifications. Chemical modifications alter the inherent features of the original nucleotides, including base-pairing performance in reverse transcription, chemical reaction activities, enzymatic reaction activities and binding affinities with specific proteins or antibodies. Therefore, coupled with next-generation sequencing, these properties of modified nucleotides can be used to characterize RNA modifications throughout the transcriptome. The existing sequencing technologies can be categorized according to the detection principles into four classes: (1) direct sequencing technology, (2) chemical-assisted sequencing technology, (3) antibody-based sequencing technology, and (4) enzyme/protein-assisted sequencing technology (Table 1).
Direct sequencing technology
For some modified nucleotides, the existence of chemical modifications alters canonical Watson-Crick base pairing during reverse transcription and further leads to truncation or misincorporation in cDNA synthesis. Therefore, this feature can be used to map modified nucleotides throughout the transcriptome.
For instance, in contrast to adenosine, inosine pairs with cytidine in reverse transcription, and thus, the A-to-I editing position can be identified by comparing the genomic DNA and RNA sequencing data and detecting A-to-G mismatch sites (Fig. 3a)111,112,113,114. Although this approach is widely applied to A-to-I editing site detection, caution is still needed to reduce the false positives introduced by SNPs, somatic mutations, pseudogenes and sequencing errors22.
a Detection of A-to-I editing sites by detecting A-to-G mismatches between genomic DNA and RNA sequencing (RNA-seq) data from the same individuals. b Detection of modified nucleotides by utilizing demethylase treatment, altered RT conditions, engineered reverse transcriptase, or modified dNTPs to generate distinct RT signatures at modified sites between treated and untreated samples. Modifications that can be detected by the corresponding approaches are listed below.
Aside from inosine, several other chemical modifications at the Watson-Crick face of the nucleobase can also induce RT stops or mutations in cDNA synthesis, thereby allowing base-resolution detection115. In contrast to inosine, the reverse transcription signatures of these modifications are not constant and dependent on both the surrounding sequences and the RT conditions116. Furthermore, to increase the detection accuracy, several improvements have been made. For instance, the E. coli AlkB demethylase and its mutant treatment are introduced to remove m1A, m3C, m1G, N2,N2-dimethyl-guanosine (m22G), and 3-methyluridine (m3U) in RNA prior to cDNA synthesis, and high-confidence methylation sites in the tRNA transcriptome can thus be identified by comparing the data for parallel sequenced, demethylase-treated and untreated samples (Fig. 3b)117,118,119,120. Moreover, the RT signatures vary substantially under different reverse transcription conditions42,121. Hence, to improve detection sensitivity, an HIV-1 reverse transcriptase against m1A was developed that allows m1A detection in human mRNA122. Moreover, RT systems, including enzymes and reaction conditions, can be optimized to detect modified nucleotides that do not interfere with Watson-Crick base pairing. For instance, Nm, an RT-silent modification, can block cDNA synthesis when the concentration of dNTPs or Mg2+ in RT reactions is limited. Coupling this feature with next-generation sequencing, two methods, 2OMe-seq and MeTH-seq, have achieved a transcriptomic profile of Nm at base resolution (Fig. 3b)123,124. As mentioned above, some DNA polymerases also possess reverse transcriptase activity and show differential elongation ability when encountering m6A residues versus A residues. To advance this feature, the KlenTaq DNA polymerase was evolved and the evolved variant exhibited significantly increased error rates opposite m6A but not unmodified A, enabling direct identification of m6A by analyzing the mutational signal from sequencing data (Fig. 3b)125. In addition, substitution of 4SedTTP (atom-specific replacement of oxygen with selenium at the 4-position) for dTTP under RT conditions can also facilitate m6A detection. In principle, compared with 4SeT-A, 4SeT-m6A pairing is unfavorable and results in aborted cDNA synthesis opposite m6A sites, thereby allowing for the discrimination of m6A from A. Furthermore, with the assistance of the m6A demethylase FTO through high-throughput sequencing, m6A can be precisely identified within the mammalian transcriptome at single-nucleotide resolution (Fig. 3b)126.
Chemical-assisted sequencing technologies
Chemical treatments are widely exploited to discriminate modified nucleotides from unmodified nucleotides in three ways: (1) installing biotin tags to enrich modified transcripts; (2) altering the base-pairing features to induce misincorporation or truncation in reverse transcription; and (3) chemical-induced cleavage followed by specific adaptor ligation (Fig. 4).
The enrichment strategy facilities RNA modification detection in low-abundance RNA species and low-stoichiometry modifications. Some modified nucleotides can be labeled by specific chemical reactions and further conjugated with the biotin molecules, thus enabling streptavidin enrichment. For instance, in borohydride reduction sequencing (BoRed-seq), total RNA is treated with NaBH4 and subsequently exposed to low pH to generate abasic sites at m7G positions, which can be tagged with biotin molecules for further streptavidin pulldown (Fig. 4a)53. Direct m6A labeling is difficult due to the inert reactivity of the methyl group. To solve this challenge, researchers developed a method named m6A-SEAL-Seq (an FTO-assisted m6A selective chemical labeling method). In this method, m6A is first oxidized to hm6A by the demethylase FTO, and hm6A is further converted to N6-dithiolsitolmethyladenosine (dm6A) through DTT-mediated thiol addition. The free sulfhydryl group of dm6A enables biotin to be conjugated to the m6A-modified transcripts, thereby facilitating streptavidin enrichment and sequencing (Fig. 4a)127.
The detection strategy of altering the base-pairing properties of nucleotides by chemical treatment has been widely applied to the transcriptome-wide detection of various modifications, including inosine, m5C, Ψ, m7G, ac4C, D and m6A. For inosine detection, the direct sequencing approach is disturbed by background noise. To overcome this limitation, a selective chemical labeling reaction with acrylonitrile is adopted, and the formed N1-cyanoethylinosine (ce1I) blocks reverse transcription, resulting in truncation of cDNA. Coupling this chemical reaction with sequencing, referred to as inosine chemical erasing sequencing (ICE-seq), can achieve base-resolution inosine detection throughout the transcriptome (Fig. 4a)128. Bisulfite treatment selectively converts unmethylated cytidine into uridine, leading to a C-to-T transition in the sequencing data; thus, transcriptome-wide m5C can be characterized by detecting nonconverted Cs in the sequencing data (Fig. 4a)129,130. Furthermore, to reduce false positives caused by insufficient conversion, several improvements have been exploited, including optimizing bisulfite treatment conditions to increase deamination efficiency131,132, using ACT random hexamers devoid of Gs to avoid copying inefficiently deaminated RNA templates16, and developing robust computational pipelines to accurately identify m5C sites132,133. Ψ can be labeled by CMC at the Watson-Crick face, and the CMC-Ψ adducts stall reverse transcription, thus inducing truncations in cDNA synthesis. Combining this chemical reaction with next-generation sequencing, researchers developed Ψ-Seq, Pseudo-seq and PSI-seq, achieving base-resolution pseudouridylation detection in yeast and mammalian transcriptomes134,135,136. To improve the robustness of Ψ detection, the chemical reaction is adapted by using a synthesized CMC derivative, azido-CMC (N3-CMC), instead of CMC. The presence of an azido group enables conjugation with biotin through a click reaction. Hence, the Ψ-containing RNA can be pre-enriched before sequencing, and this method is named CeU-seq86. The pre-enrichment step enables CeU-seq to identify thousands of Ψ sites in the mammalian transcriptome (Fig. 4a). In addition to CMC labeling, a recent work showed that Ψ can form a stable monobisulfite adduct upon bisulfite treatment and further leave a deletion signature at the exact modified sites, thereby providing an orthogonal Ψ detection strategy (Fig. 4a)131. As described above, m7G residues can be converted to abasic sites upon NaBH4 treatment and further recorded as misincorporations through reverse transcription and sequencing. Accordingly, the m7G Mutational Profiling sequencing (m7G-MaP-seq) can map internal m7G modifications at nucleotide resolution in tRNA and rRNA137. To achieve m7G detection in mRNA, m7G-seq conjugates a biotin molecule to the generated abasic sites, thus enabling pre-enrichment of m7G-containing RNA. Moreover, the biotinylated sites induce misincorporations during cDNA synthesis; therefore, transcriptome-wide base-resolution m7G mapping can be achieved (Fig. 4a)54. In addition, D can also be reduced by NaBH4 treatment, and the reduction product can be further labeled by Rho (rhodamine). Furthermore, the Rho-adducts block reverse transcriptase elongation and thus can be identified by analyzing induced RT-stops in sequencing data (Fig. 4a)138. ac4C can react with NaCNBH3 under acidic conditions, and the formed reduced nucleobase, N4-acetyltetrahydrocytidine, causes misincorporation during cDNA synthesis. Taking advantage of this reaction, ac4C-seq can map ac4C at single-nucleotide resolution (Fig. 4a)139. In addition to altering the structure of nucleotides by chemical reactions, the chemical group can also be introduced through metabolic labeling. For instance, when feeding cells with an S-adenosyl methionine (SAM) analog, Se-allyl-L-selenohomocysteine, the cellular RNAs could be modified with N6-allyladenosine (a6A) at supposed m6A-generating sites. Furthermore, a6A-containing RNAs can be enriched by a specific antibody, and a6A sites are converted to N1,N6-cyclized adenosine (cyc-A) through the iodination-induced cyclization reaction. As cyc-A induces misincorporations in cDNA synthesis, m6A can be mapped at base resolution by detecting mutation signals in sequencing data (Fig. 4b)140. With the exception of metabolic labeling, the allyl group can also be transferred to m6A by the Dim1/KsgA family of dimethyltransferases, which can specifically convert m6A into allyl-modified m6A (N6-allyl, N6-methyladenosine, a6m6A). Since cyclized a6m6A can induce misincorporation in RT, the technology, named m6A-SAC-seq, achieves quantitative, transcriptome-wide mapping of m6A at single nucleotide resolution141.
Compared with unmodified nucleotides, modified nucleotides exhibit distinct resistance under chemical hydrolysis treatment. Based on this principle, several sequencing methods have been developed and have achieved transcriptome-wide profiling for Nm, m7G, m3C and Ψ. Nm is resistant to alkaline hydrolysis and thus can be mapped at single-base resolution by analyzing read-ends information in sequencing data. Since RNA fragmentation is random and irregular, this strategy requires rather high read coverage and is limited to highly abundant RNAs142,143,144. To overcome this limitation, ribose oxidation sequencing (RibOxi-seq) and Nm-seq have been developed. In these two methods, RNA is treated with iterative oxidation–elimination–dephosphorylation (OED) cycles to remove unmodified nucleotides, and the non-methylated ends cannot be ligated to linkers for sequencing library construction145,146. The selective cleavage and ligation allow detection of low-stoichiometry 2ʹ-O-methylation sites using this approach. Similarly, m7G and m3C are resistant to NaBH4-aniline treatment, and the generated 5′-phosphate end during aniline cleavage could be exploited for selective ligation to enrich modified fragments147,148. This positive selection strategy facilitates transcriptome-wide detection of m7G and m3C. In addition, upon hydrazine-aniline treatment, RNA can be specifically cleaved at m3C-modified sites and resistant to cleavage at Ψ-modified sites. Based on this approach, hydrazine-aniline cleavage sequencing (HAC-seq) and HydraPsiSeq have been developed to detect m3C and Ψ, respectively (Fig. 4a)149,150.
Antibody-based sequencing technologies
Antibody-based strategies have been exploited for transcriptome-wide mapping of several RNA modifications, including m6A/m6Am, m1A, hm5C, ac4C and m7G (Fig. 5)39,53,54,55,148,151,152,153,154. In this strategy, isolated RNA is first fragmented to 100–200 nt, and certain modification-containing RNA fragments are enriched by specific antibody immunoprecipitation. The enriched RNAs are subjected to high-throughput sequencing, and the modifications of interest can be identified by bioinformatic analysis. Robust enrichment allows antibody-based strategies to be very sensitive in detecting low-abundance modifications in mRNA and other rare RNA species.
However, limited by the fragmentation size, the resolution of the antibody-based strategy is approximately 100–200 bp. To improve the detection resolution, several improvements have been made. For instance, introducing UV-induced RNA-antibody crosslinking can lead to truncation or misincorporation at protein–RNA crosslinking sites during reverse transcription, thereby allowing transcriptome-wide single-base resolution detection for m6A and m7G, respectively (Fig. 5a)155,156,157. In addition, coupling antibody immunoprecipitation with enzyme/chemical treatment can not only increase detection resolution but also reduce false positives. For example, taking advantage of the fact that the m1A-induced mutational RT signatures can be erased by AlkB demethylase treatment or Dimroth rearrangement, m1A-MAP, m1A-seq and m1A-IP-Seq have achieved base-resolution m1A methylome detection (Fig. 5a)42,122,158. In addition, utilizing selective in vitro demethylation for m6Am, m6Am-seq has the capability of discriminating m6Am from m6A and can identify m6Am at base resolution (Fig. 5b)159.
Enzyme/protein-assisted sequencing technologies
In addition to antibody immunoprecipitation, some enzymes or RNA modification-related proteins can also be utilized for affinity capture or editing modification-containing transcripts, thereby enabling transcriptomic RNA modification detection (Fig. 6). For instance, by feeding a 5-aza-C analog or overexpressing mutated Nsun2, a covalent bond can be formed between m5C methyltransferase and its target sites. Based on this approach, Aza-IP (5-azacytidine–mediated RNA immunoprecipitation) and miCLIP (methylation iCLIP) can enrich target sites by immunoprecipitation, thereby enabling identification of the direct targets of m5C methyltransferases (Fig. 6a)160,161. In addition to methyltransferase, reader proteins can also be used to target modified nucleotides. For example, in DART-seq (deamination adjacent to RNA modification targets), the cytidine deaminase APOBEC1 is fused to the m6A-binding YTH domain and thus leads to C-to-U deamination at sites adjacent to m6A residues. Furthermore, m6A residues can be identified by analyzing C-to-T mismatches in sequencing data (Fig. 6b)162. Recently, the authors further integrated DART-seq with a single-cell RNA-sequencing platform and thus developed scDART-seq, achieving profiling of the m6A methylome in single cells163. In addition, some RNA exonuclease also possesses binding affinity for certain RNA modifications under certain conditions. For instance, in the presence of Ca2+, E. coli Endonuclease V (eEndoV) promotes binding of inosine in RNA instead of cleavage. Taking advantage of this approach, Endonuclease V immunoprecipitation enrichment sequencing (EndoVIPER-seq) can enrich A-to-I edited transcripts from cellular RNA164.
Similar to chemical-induced hydrolysis, some endonucleases also show distinct cleavage efficiency between modified and unmodified transcripts, which can be utilized to enrich modification-containing transcripts and identify modified nucleotides. For instance, the E. coli RNA endoribonuclease MazF can specifically cleave the unmethylated 5ʹ-ACA-3ʹ motif but not the 5ʹ-m6ACA-3ʹ motif165. Taking advantage of this specificity, RNA-endoribonuclease–facilitated sequencing (m6A–REF-seq)/MAZTER-seq allows quantitative profiling of m6A at single-nucleotide resolution (Fig. 6c)166,167. However, a major limitation of this approach is that it can only detect m6A in the ACA context, which is only a small portion (16-25%) of m6A-modified sites.
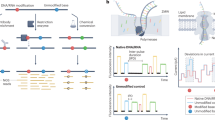
Nanopore direct RNA sequencing-based detection technology
Next-generation sequencing-based detection technologies have been widely applied for transcriptome-wide RNA modification detection. However, limited by the sequencing length (from 50 to 300 bp) and distinct detection principles for different modifications, next-generation sequencing-based detection technologies cannot map diverse RNA modifications simultaneously. The development of the Oxford Nanopore Technologies (ONT) sequencing platform shows promise in overcoming these challenges. In contrast to next-generation RNA sequencing, nanopore sequencing can sequence RNA directly without the requirement of additional reverse transcription and PCR amplification, thus decreasing the biases caused by these steps168,169,170,171. Mechanistically, single-stranded RNA is driven through the nanopore by the motor protein and thus causes ionic current changes for a set of k nucleotides residing within the pore (kmer; typically, k is 5), which enables decoding of the nucleotide sequence by computational analysis (Fig. 7). In addition to sequence information, chemical modifications and secondary structure, which also influence RNA translocation in nanopores, can be determined directly by computational algorithms172. In addition, the reads generated by nanopore sequencing are long enough to cover the full length of a transcript, thereby enabling accurate identification of highly repetitive regions, spliced products and polyadenylation tail length.
To identify RNA modification from the nanopore RNA direction, robust computational algorithm analysis is vital. There are two major analysis strategies, including alterations of raw signal intensity analysis (signal intensity, dwell time and trace) and base-called “error” feature analysis (base quality, mismatch frequency, and deletion frequency)170. Furthermore, modifications can be identified by the algorithms previously trained with modified and unmodified kmer contexts or comparison with the nonmodified control samples. These computational algorithms have allowed the identification of several modifications, including m6A, m7G, m5C, hm5C, Ψ and Nm, using nanopore direct RNA sequencing169,172,173,174,175,176,177,178,179,180. In addition, owing to the long sequencing length, the sequence information and modification landscape of SARS-CoV-2 can be determined simultaneously by nanopore direct RNA sequencing181,182. However, there are still many challenges and limitations for nanopore RNA sequencing-based detection technology. First, the sequencing error rate (~1–4%)168 is still much higher than that of next-generation sequencing (~0.1-1%). Since some algorithms exploit systematic base-calling errors to identify RNA modifications, the high sequencing error rate increases the complexity for base calling and modification identification. Second, the throughput of nanopore RNA sequencing is relatively low (1–3 Gb per flow cell), and the costs of direct RNA sequencing are high. Recently, several studies have shown that accurate modification detection requires high sequencing coverage (at least 30X)173,180. Hence, the sequencing depth and costs limit broader application of this strategy. Third, nanopore direct RNA sequencing requires a large amount of RNA (~500 ng polyA+ RNA), and thus, it is not suitable for samples that are difficult to obtain. Therefore, efforts are still needed to improve nanopore RNA direct sequencing, including the development of more powerful nanopore proteins and robust computational algorithms.
Discussion
Since all existing detection methods confer both advantages and drawbacks, appropriate detection strategies should be chosen by considering the abundance of RNA modifications, RNA types of interest (rRNA, tRNA, mRNA or miRNA), the amount of starting material, etc. For instance, direct sequencing methods preserve the modification status but are often confounded by other factors, including RNA secondary structure, SNPs, and sequencing errors. Direct sequencing methods also require a high sequencing depth to detect modifications in low-abundance RNA. Antibody-based detection technologies are powerful and capable of detecting low-abundance modifications in rare RNA species. However, due to the enrichment step, the stoichiometry information of modification is lost, and large starting materials are needed. In addition, since antibodies may have intrinsic bias on RNA sequences and secondary structures, caution is needed to eliminate false-positive signals. Chemical treatment-based approaches have been widely applied to RNA modification detection owing to their high labeling specificity. For this strategy, both high labeling efficiency and mild reaction conditions are required for transcriptome-wide modification detection and quantification. Metabolic labeling or RNA modification-related protein-based approaches provide an alternative strategy to characterize modified residues but can only be applied to cultured cells. Therefore, appropriate negative controls are necessary to reduce false positives, including modification status manipulation (enzyme knockdown/knockout), chemical or enzyme treatment, and differential RT conditions. Orthogonal locus-specific detection methods are also necessary to validate the modification sites and status. In addition, synthetic modified spike-in controls preserve stoichiometry information during sequencing122,183. In addition, synthetic RNAs can also be developed for reducing false positives in sequencing-based technologies. For instance, an elegant study, which utilizes an unmodified IVT RNA library resembling the endogenous transcriptome as a negative control, can precisely and quantitatively map m6A and m5C through calibration184.
Nanopore direct RNA sequencing has been applied to the detection of several RNA modifications, showing promise for the simultaneous identification of distinct modifications in a single molecule. However, the accuracy and sensitivity of nanopore direct RNA modification sequencing are limited, and the simultaneous detection of different modifications is still unachieved. Further efforts should be made to improve the instrument to reduce the signal noise and develop robust computational analysis to identify multiple modifications simultaneously. Finally, the development of RNA modification detection technologies for low-input and single-cell samples is still an urgent need for facilitating understanding of the roles of RNA modifications in physiological and pathological processes. Improvement in integrating existing RNA modification detection strategies with single-cell RNA sequencing platforms shows promise. Collectively, recent progress in detection technologies promotes functional studies of RNA modifications, and we anticipate that further advancement will lead to a more comprehensive understanding of the epitranscriptome.
References
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Wang, Y. & Jia, G. Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 64, 967–979 (2020).
Wang, Y., Zhang, X., Liu, H. & Zhou, X. Chemical methods and advanced sequencing technologies for deciphering mRNA modifications. Chem. Soc. Rev. 50, 13481–13497 (2021).
Wiener, D. & Schwartz, S. The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 22, 119–131 (2021).
Owens, M. C., Zhang, C. & Liu, K. F. Recent technical advances in the study of nucleic acid modifications. Mol. Cell 81, 4116–4136 (2021).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Yang, Y., Hsu, P. J., Chen, Y. S. & Yang, Y. G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 28, 616–624 (2018).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Delaunay, S. & Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 21, 552–559 (2019).
Chen, Y. S., Yang, W. L., Zhao, Y. L. & Yang, Y. G. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 12, e1639 (2021).
Shanmugam, R. et al. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Disco. 1, 15010 (2015).
Shen, H. et al. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem. 296, 100087 (2021).
He, C. et al. TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol. 28, 62–70 (2021).
Yang, X. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 27, 606–625 (2017).
Chen, X. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019).
Yang, Y. et al. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 75, 1188–-1202.e1111 (2019).
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014).
Gerber, A. P. & Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149 (1999).
Seeburg, P. H. & Hartner, J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 13, 279–283 (2003).
Wulff, B. E., Sakurai, M. & Nishikura, K. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat. Rev. Genet. 12, 81–85 (2011).
Arnez, J. G. & Steitz, T. A. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33, 7560–7567 (1994).
Karijolich, J., Yi, C. & Yu, Y. T. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 16, 581–585 (2015).
Newby, M. I. & Greenbaum, N. L. Investigation of overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl Acad. Sci. USA 99, 12697–12702 (2002).
Davis, D. R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23, 5020–5026 (1995).
King, T. H., Liu, B., McCully, R. R. & Fournier, M. J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11, 425–435 (2003).
Liang, X. H., Liu, Q. & Fournier, M. J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 28, 965–977 (2007).
Decatur, W. A. & Fournier, M. J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
Baudin-Baillieu, A. et al. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37, 7665–7677 (2009).
Jack, K. et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44, 660–666 (2011).
Li, X., Ma, S. & Yi, C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 33, 108–116 (2016).
Kariko, K. et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008).
Eyler, D. E. et al. Pseudouridinylation of mRNA coding sequences alters translation. Proc. Natl Acad. Sci. USA 116, 23068–23074 (2019).
Karijolich, J. & Yu, Y. T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011).
Schevitz, R. W. et al. Crystal structure of a eukaryotic initiator tRNA. Nature 278, 188–190 (1979).
Helm, M. et al. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 26, 1636–1643 (1998).
Voigts-Hoffmann, F. et al. A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys). J. Am. Chem. Soc. 129, 13382–13383 (2007).
Dominissini, D. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Ballesta, J. P. & Cundliffe, E. Site-specific methylation of 16S rRNA caused by pct, a pactamycin resistance determinant from the producing organism, Streptomyces pactum. J. Bacteriol. 173, 7213–7218 (1991).
Waku, T. et al. NML-mediated rRNA base methylation links ribosomal subunit formation to cell proliferation in a p53-dependent manner. J. Cell Sci. 129, 2382–2393 (2016).
Li, X. et al. Base-Resolution Mapping Reveals Distinct m(1)A Methylome in Nuclear- and Mitochondrial-Encoded Transcripts. Mol. Cell 68, 993–1005.e1009 (2017).
Saikia, M., Fu, Y., Pavon-Eternod, M., He, C. & Pan, T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16, 1317–1327 (2010).
Chan, C. T. et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6, e1001247 (2010).
Helm, M. & Alfonzo, J. D. Posttranscriptional RNA Modifications: playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 21, 174–185 (2014).
Peifer, C. et al. Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res. 41, 1151–1163 (2013).
Xiong, X., Li, X. & Yi, C. N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 45, 179–186 (2018).
Elliott, B. A. et al. Modification of messenger RNA by 2’-O-methylation regulates gene expression in vivo. Nat. Commun. 10, 3401 (2019).
Choi, J. et al. 2’-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 25, 208–216 (2018).
Hoernes, T. P. et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 852–862 (2016).
Hoernes, T. P. et al. Eukaryotic Translation Elongation is Modulated by Single Natural Nucleotide Derivatives in the Coding Sequences of mRNAs. Genes (Basel) 10, 84 (2019).
Ayadi, L., Galvanin, A., Pichot, F., Marchand, V. & Motorin, Y. RNA ribose methylation (2’-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 253–269 (2019).
Pandolfini, L. et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 74, 1278–1290.e1279 (2019).
Zhang, L. S. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell 74, 1304–1316.e1308 (2019).
Arango, D. et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell 175, 1872–1886.e1824 (2018).
Kumbhar, B. V., Kamble, A. D. & Sonawane, K. D. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 66, 797–816 (2013).
Ito, S. et al. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J. Biol. Chem. 289, 26201–26212 (2014).
Jin, G., Xu, M., Zou, M. & Duan, S. The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: a systematic review. Mol. Ther. Nucleic Acids 20, 13–24 (2020).
Dominissini, D. & Rechavi, G. N(4)-acetylation of Cytidine in mRNA by NAT10 Regulates Stability and Translation. Cell 175, 1725–1727 (2018).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA Biol. 7, 237–247 (2010).
Stanley, J. & Vassilenko, S. A different approach to RNA sequencing. Nature 274, 87–89 (1978).
Gupta, R. C. & Randerath, K. Rapid print-readout technique for sequencing of RNA’s containing modified nucleotides. Nucleic Acids Res. 6, 3443–3458 (1979).
Keith, G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 77, 142–144 (1995).
Zhao, X. & Yu, Y. T. Detection and quantitation of RNA base modifications. RNA 10, 996–1002 (2004).
Grosjean, H., Droogmans, L., Roovers, M. & Keith, G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 425, 55–101 (2007).
Grosjean, H., Keith, G. & Droogmans, L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 265, 357–391 (2004).
Jia, G. F. et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO (vol 7, pg 885, 2011). Nat. Chem. Biol. 8, 1008–1008 (2012).
Nagarajan, A., Janostiak, R. & Wajapeyee, N. Dot Blot Analysis for Measuring Global N(6)-Methyladenosine Modification of RNA. Methods Mol. Biol. 1870, 263–271 (2019).
Nees, G., Kaufmann, A. & Bauer, S. Detection of RNA modifications by HPLC analysis and competitive ELISA. Methods Mol. Biol. 1169, 3–14 (2014).
Yoluc, Y. et al. Instrumental analysis of RNA modifications. Crit. Rev. Biochem. Mol. Biol. 56, 178–204 (2021).
Gaston, K. W. & Limbach, P. A. The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry. RNA Biol. 11, 1568–1585 (2014).
Kellner, S. et al. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 42, e142 (2014).
Thuring, K., Schmid, K., Keller, P. & Helm, M. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods 107, 48–56 (2016).
Bar-Yaacov, D. et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 14, e1002557 (2016).
Sharma, S. et al. A single N(1)-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci. Rep. 8, 11904 (2018).
Woo, H. H. & Chambers, S. K. Human ALKBH3-induced m(1)A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 35–46 (2019).
Jackman, J. E., Montange, R. K., Malik, H. S. & Phizicky, E. M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9, 574–585 (2003).
Lee, C., Kramer, G., Graham, D. E. & Appling, D. R. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J. Biol. Chem. 282, 27744–27753 (2007).
Bakin, A. & Ofengand, J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32, 9754–9762 (1993).
Datta, B. & Weiner, A. M. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature 352, 821–824 (1991).
Herschlag, D. Biophysical, chemical, and functional probes of RNA structure, interactions and folding: Part A. Preface. Methods Enzymol. 468, xv (2009).
Maden, B. E., Corbett, M. E., Heeney, P. A., Pugh, K. & Ajuh, P. M. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie 77, 22–29 (1995).
Kiss, T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002).
Yu, Y. T., Shu, M. D. & Steitz, J. A. A new method for detecting sites of 2’-O-methylation in RNA molecules. RNA 3, 324–331 (1997).
Liu, N. et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856 (2013).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Dong, Z. W. et al. RTL-P: a sensitive approach for detecting sites of 2’-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Aschenbrenner, J. & Marx, A. Direct and site-specific quantification of RNA 2’-O-methylation by PCR with an engineered DNA polymerase. Nucleic Acids Res. 44, 3495–3502 (2016).
Harcourt, E. M., Ehrenschwender, T., Batista, P. J., Chang, H. Y. & Kool, E. T. Identification of a selective polymerase enables detection of N(6)-methyladenosine in RNA. J. Am. Chem. Soc. 135, 19079–19082 (2013).
Castellanos-Rubio, A. et al. A novel RT-QPCR-based assay for the relative quantification of residue specific m6A RNA methylation. Sci. Rep. 9, 4220 (2019).
Wang, S. et al. N(6)-Methyladenine hinders RNA- and DNA-directed DNA synthesis: application in human rRNA methylation analysis of clinical specimens. Chem. Sci. 7, 1440–1446 (2016).
Xiao, Y. et al. An elongation- and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-specific N(6) -methyladenosine modification. Angew. Chem. Int. Ed. Engl. 57, 15995–16000 (2018).
Wang, Y. et al. LEAD-m(6) A-seq for Locus-Specific Detection of N(6) -Methyladenosine and Quantification of Differential Methylation. Angew. Chem. Int. Ed. Engl. 60, 873–880 (2021).
Qu, H. L., Michot, B. & Bachellerie, J. P. Improved methods for structure probing in large RNAs: a rapid ‘heterologous’ sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5’ terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 11, 5903–5920 (1983).
Lei, Z. & Yi, C. A Radiolabeling-Free, qPCR-Based Method for Locus-Specific Pseudouridine Detection. Angew. Chem. Int. Ed. Engl. 56, 14878–14882 (2017).
Schurch, S. Characterization of nucleic acids by tandem mass spectrometry - The second decade (2004-2013): From DNA to RNA and modified sequences. Mass Spectrom. Rev. 35, 483–523 (2016).
Jora, M., Lobue, P. A., Ross, R. L., Williams, B. & Addepalli, B. Detection of ribonucleoside modifications by liquid chromatography coupled with mass spectrometry. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 280–290 (2019).
Yuan, B. F. Liquid chromatography-mass spectrometry for analysis of RNA adenosine methylation. Methods Mol. Biol. 1562, 33–42 (2017).
Suzuki, T., Ikeuchi, Y., Noma, A., Suzuki, T. & Sakaguchi, Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 425, 211–229 (2007).
Akichika, S., Suzuki, T. & Suzuki, T. Mass spectrometric analysis of mRNA 5’ terminal modifications. Methods Enzymol. 658, 407–418 (2021).
Kurata, T. et al. RelA-SpoT Homolog toxins pyrophosphorylate the CCA end of tRNA to inhibit protein synthesis. Mol. Cell 81, 3160–3170.e3169 (2021).
Bangs, J. D., Crain, P. F., Hashizume, T., McCloskey, J. A. & Boothroyd, J. C. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267, 9805–9815 (1992).
Guymon, R., Pomerantz, S. C., Crain, P. F. & McCloskey, J. A. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry 45, 4888–4899 (2006).
Guymon, R., Pomerantz, S. C., Ison, J. N., Crain, P. F. & McCloskey, J. A. Post-transcriptional modifications in the small subunit ribosomal RNA from Thermotoga maritima, including presence of a novel modified cytidine. RNA 13, 396–403 (2007).
Puri, P. et al. Systematic identification of tRNAome and its dynamics in Lactococcus lactis. Mol. Microbiol. 93, 944–956 (2014).
Suzuki, T. & Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 42, 7346–7357 (2014).
Mandal, D. et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl Acad. Sci. USA 107, 2872–2877 (2010).
Miyauchi, K., Kimura, S. & Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 9, 105–111 (2013).
Kang, B. I. et al. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 45, 2124–2136 (2017).
Akichika, S. et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Ju, Y. S. et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat. Genet. 43, 745–752 (2011).
Bahn, J. H. et al. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22, 142–150 (2012).
Ramaswami, G. et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods 9, 579–581 (2012).
Peng, Z. et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30, 253–260 (2012).
Ryvkin, P. et al. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692 (2013).
Hauenschild, R. et al. The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res. 43, 9950–9964 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22, 1771–1784 (2016).
Dai, Q., Zheng, G., Schwartz, M. H., Clark, W. C. & Pan, T. Selective Enzymatic Demethylation of N(2),N(2) -Dimethylguanosine in RNA and Its Application in High-Throughput tRNA Sequencing. Angew. Chem. Int. Ed. Engl. 56, 5017–5020 (2017).
Werner, S. et al. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res. 48, 3734–3746 (2020).
Zhou, H. et al. Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods 16, 1281–1288 (2019).
Incarnato, D. et al. High-throughput single-base resolution mapping of RNA 2-O-methylated residues. Nucleic Acids Res. 45, 1433–1441 (2017).
Bartoli, K. M., Schaening, C., Carlile, T. M. & Gilbert, W. V. Conserved Methyltransferase Spb1 Targets mRNAs for Regulated Modification with 2′-O-Methyl Ribose. Preprint at https://www.biorxiv.org/content/10.1101/271916v2 (2018).
Aschenbrenner, J. et al. Engineering of a DNA Polymerase for Direct m(6) A Sequencing. Angew. Chem. Int. Ed. Engl. 57, 417–421 (2018).
Hong, T. et al. Precise Antibody-Independent m6A Identification via 4SedTTP-Involved and FTO-Assisted Strategy at Single-Nucleotide Resolution. J. Am. Chem. Soc. 140, 5886–5889 (2018).
Wang, Y., Xiao, Y., Dong, S., Yu, Q. & Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat. Chem. Biol. 16, 896–903 (2020).
Sakurai, M. et al. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 24, 522–534 (2014).
Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9, e1003602 (2013).
Khoddami, V. et al. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl Acad. Sci. USA 116, 6784–6789 (2019).
Huang, T., Chen, W., Liu, J., Gu, N. & Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 26, 380–388 (2019).
Legrand, C. et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27, 1589–1596 (2017).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Enroth, C. et al. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 47, e126 (2019).
Finet, O. et al. Transcription-wide mapping of dihydrouridine reveals that mRNA dihydrouridylation is required for meiotic chromosome segregation. Mol. Cell 82, 404–419.e9 (2021).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020).
Shu, X. et al. A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat. Chem. Biol. 16, 887–895 (2020).
Hu, L. et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 1–10 (2022).
Birkedal, U. et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. 54, 451–455 (2015).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2’-O-Me residues in RNA. Nucleic Acids Res. 44, e135 (2016).
Krogh, N. et al. Profiling of 2’-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 44, 7884–7895 (2016).
Zhu, Y., Pirnie, S. P. & Carmichael, G. G. High-throughput and site-specific identification of 2’-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 23, 1303–1314 (2017).
Dai, Q. et al. Nm-seq maps 2’-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Marchand, V. et al. AlkAniline-Seq: profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew. Chem. Int. Ed. Engl. 57, 16785–16790 (2018).
Lin, S. et al. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e245 (2018).
Cui, J., Liu, Q., Sendinc, E., Shi, Y. & Gregory, R. I. Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res. 49, e27 (2021).
Marchand, V. et al. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 48, e110 (2020).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Delatte, B. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Linder, B. et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Chen, K. et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54, 1587–1590 (2015).
Malbec, L. et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 29, 927–941 (2019).
Safra, M. et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255 (2017).
Sun, H. et al. m(6)Am-seq reveals the dynamic m(6)Am methylation in the human transcriptome. Nat. Commun. 12, 4778 (2021).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013).
Khoddami, V. & Cairns, B. R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013).
Meyer, K. D. DART-seq: an antibody-free method for global m(6)A detection. Nat. Methods 16, 1275–1280 (2019).
Tegowski, M., Flamand, M. N. & Meyer, K. D. scDART-seq reveals distinct m(6)A signatures and mRNA methylation heterogeneity in single cells. Mol. Cell 82, 868–878.e810 (2022).
Knutson, S. D., Arthur, R. A., Johnston, H. R. & Heemstra, J. M. Selective enrichment of A-to-I edited transcripts from cellular RNA using endonuclease V. J. Am. Chem. Soc. 142, 5241–5251 (2020).
Imanishi, M., Tsuji, S., Suda, A. & Futaki, S. Detection of N(6)-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. (Camb.) 53, 12930–12933 (2017).
Garcia-Campos, M. A. et al. Deciphering the “m(6)A code” via antibody-independent quantitative profiling. Cell 178, 731–747.e716 (2019).
Zhang, Z. et al. Single-base mapping of m(6)A by an antibody-independent method. Sci. Adv. 5, eaax0250 (2019).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Begik, O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 39, 1278–1291 (2021).
Furlan, M. et al. Computational methods for RNA modification detection from nanopore direct RNA sequencing data. RNA Biol. 18, 31–40 (2021).
Wan, Y. K., Hendra, C., Pratanwanich, P. N. & Goke, J. Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data. Trends Genet 38, 246–257 (2021).
Jenjaroenpun, P. et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 49, e7 (2021).
Leger, A. et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 12, 7198 (2021).
Parker, M. T. et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m(6)A modification. Elife 9, e49658 (2020).
Liu, H. et al. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Smith, A. M., Jain, M., Mulroney, L., Garalde, D. R. & Akeson, M. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One 14, e0216709 (2019).
Price, A. M. et al. Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 11, 6016 (2020).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Gao, Y. et al. Quantitative profiling of N(6)-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using nanopore direct RNA sequencing. Genome Biol. 22, 22 (2021).
Pratanwanich, P. N. et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 39, 1394–1402 (2021).
Kim, D. et al. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921 e910 (2020).
Miladi, M. et al. The landscape of SARS-CoV-2 RNA modifications. Preprint at https://www.biorxiv.org/content/10.1101/2020.07.18.204362v1 (2020).
Koh, C. W. Q., Goh, Y. T. & Goh, W. S. S. Atlas of quantitative single-base-resolution N(6)-methyl-adenine methylomes. Nat. Commun. 10, 5636 (2019).
Zhang, Z. et al. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods 18, 1213–1222 (2021).
Acknowledgements
This work was supported by National Natural Science Foundation of China (32171283 to X.L.) and Zhejiang Provincial Natural Science Foundation of China (LR22C060002 to X.L.). The authors apologize for not being able to cite all the publications related to this topic owing to space constraints of the journal.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Y., Lu, L. & Li, X. Detection technologies for RNA modifications. Exp Mol Med 54, 1601–1616 (2022). https://doi.org/10.1038/s12276-022-00821-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-022-00821-0
This article is cited by
-
Decoding epitranscriptomic regulation of viral infection: mapping of RNA N6-methyladenosine by advanced sequencing technologies
Cellular & Molecular Biology Letters (2024)
-
Epitranscriptome insights into Riccia fluitans L. (Marchantiophyta) aquatic transition using nanopore direct RNA sequencing
BMC Plant Biology (2024)
-
Isoform-specific RNA structure determination using Nano-DMS-MaP
Nature Protocols (2024)
-
Single-molecule epitranscriptomic analysis of full-length HIV-1 RNAs reveals functional roles of site-specific m6As
Nature Microbiology (2024)
-
Transfer learning enables identification of multiple types of RNA modifications using nanopore direct RNA sequencing
Nature Communications (2024)