Abstract
The concept of the antibiotic resistome was introduced just over a decade ago, and since then, active resistome studies have been conducted. In the present study, we describe the previously established concept of the resistome, which encompasses all types of antibiotic resistance genes (ARGs), and the important findings from each One-Health sector considering this concept, thereby emphasizing the significance of the One-Health approach in understanding ARG transmission. Cutting-edge research methodologies are essential for deciphering the complex resistome structure in the microbiomes of humans, animals, and the environment. Based on the recent achievements of resistome studies in multiple One-Health sectors, future directions for resistome research have been suggested to improve the understanding and control of ARG transmission: (1) ranking the critical ARGs and their hosts; (2) understanding ARG transmission at the interfaces of One-Health sectors; (3) identifying selective pressures affecting the emergence, transmission, and evolution of ARGs; and (4) elucidating the mechanisms that allow an organism to overcome taxonomic barriers in ARG transmission.
Similar content being viewed by others
Introduction
Since the discovery of the first antibiotic penicillin1, antibiotics have been considered essential drugs for treating bacterial infections; however, unlike with other medicines, antibiotic drug resistance has become a great concern owing to its continuous emergence and rapid dissemination among pathogens, as reported in recent cases, posing substantial clinical threats2,3. Since the use of early-date antibiotics, such as penicillin and salvarsan, emergence and dissemination of resistance have been observed after introducing antibiotics into clinical settings4,5; this resistance is considered to be mainly mediated by antibiotic resistance genes (ARGs). In 1973, environmental ARGs were characterized to be genetically similar to clinical ARGs, suggesting that these clinical genes originated from antibiotic-producing actinomycetes6. Consequently, after this discovery, ARGs were intensively studied in only clinical pathogens for >30 years until the resistome concept was proposed7; this concept has provided exceptional insights into the origin and dissemination of ARGs. This review aims to describe the concept of the antibiotic resistome; summarize the information from the last decade regarding approaches under the concept; and provide a comprehensive understanding of the origin, emergence, dissemination, and evolution of ARGs. In particular, resistome studies from the One-Health (Human–Animal–Environment) perspective will be essential for deciphering the complex resistome structure and determining prioritized factors to aid in the mitigation of ARG transmission to the clinic.
Concept of the antibiotic resistome
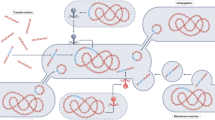
The term “antibiotic resistome” was first coined in 2006 by Gerry Wright’s group, who defined the soil resistome as “resistance determinants present in the soil”, thereby demonstrating that multidrug resistance in a population of environmental bacteria was more prevalent than previously assumed7. Later, the resistome was defined as “a collection of all the ARGs and their precursors in pathogenic and nonpathogenic bacteria”8. Its constituents were precisely described as “all ARGs, including those circulating in pathogenic bacteria, antibiotic producers, and benign nonpathogenic bacteria”9. Considering the origin of ARGs, the resistome has been suggested to include protoresistance genes as a deep reservoir of ARG precursors, as well as clinical, environmental, and intrinsic resistance genes10. After a large number of studies on the resistome were published, the definition was further refined by designating types of resistance, such as acquired resistance (vertically or horizontally transferred, taxa-nonspecific), intrinsic resistance (only vertically transmitted, taxa-specific), silent/cryptic resistance (phenotypically sensitive, functional but not expressed), and protoresistance (phenotypically sensitive, little/no activity until mutated)11. In summary, the antibiotic resistome encompasses all types of ARGs (acquired and intrinsic resistance genes), their precursors, and some potential resistance mechanisms within microbial communities that require evolution or alterations in the expression context to confer resistance (Fig. 1).
Since 2006, critical information about antimicrobial resistance (AMR) has been revealed based on the resistome concept. Old speculations were confirmed, and critical new findings were suggested as follows: (i) AMR is ancient and ubiquitous in various microbiomes12,13,14; (ii) the antibiotic resistome is complex and diverse15,16; (iii) the environmental resistome is the origin and a reservoir of ARGs13,15,17; (iv) the resistome is generally determined by the microbial community structure in the natural environment18; (v) anthropogenic activities shape the environmental resistome19; (vi) mobile genetic elements are responsible for ARG transmission20; and (vii) ARGs flow among humans, animals, and the environment21. These discoveries have provided the base for recent approaches aimed at improving the understanding of AMR in the human, animal, and environmental sectors, namely, the “One-Health” approach.
Antibiotic resistome in One-Health sectors
The One-Health concept
As mentioned above, ARGs circulate among the microbiomes of humans, animals, and the environment, which form the different sectors of the One-Health concept. One-Health is defined as the “collaborative effort of multiple disciplines–working locally, nationally, and globally–to attain optimal health for people, animals, and the environment through policy, research, education, and practice”22,23. Initially, zoonoses were recognized as major issues in both humans and animals. In 2008, the importance of “ecosystems” was particularly emphasized in the One-Health concept; this was documented by consultation from several organizations including the Food and Agriculture Organization (FAO), World Organization for Animal Health (OIE), and World Health Organization (WHO) (http://www.fao.org/3/aj137e/aj137e00.htm). In general, infectious diseases, including zoonoses, are the main concerns and targets of the One-Health approach. AMR, which is directly related to such diseases, was considered one of the three One-Health priorities during the tripartite (FAO-OIE-WHO) meeting in 201124. Considering ARG flow among the One-Health sectors, it has been recognized as one of the main issues in the One-Health approach25,26. The One-Health concept has focused on interconnections and interdependencies among sectors at local sites; however, recently, considering global health, the comprehension of communication among local ecosystems and the identification of factors that stimulate the global AMR crisis has also gained immense attention27. In the present study, we review the achievements in each sector and evaluate the factors that need to be prioritized to mitigate ARG transmission to the clinic.
Environmental resistome
Since the resistome concept originated in the soil environment, the environmental resistome has been extensively deciphered7. ARGs, as ancient and ubiquitous components of bacteria, exist in all ecological niches that harbor various microbial communities13,28. From the viewpoint of microbial ecology, the environment can be classified into natural and built environments, and the former can be divided into aquatic and terrestrial environments. The aquatic environment includes marine ecosystems (oceans and estuaries) and freshwater ecosystems (rivers, lakes, and wetlands). The terrestrial environment includes all kinds of terrestrial ecosystems in various climate zones such as forests, deserts, grasslands, and tundra. Built environments include wastewater treatment plants (WWTPs), agricultural sites, aquaculture operations, and hospital environments. In this review, we focus on the environmental resistomes in soil, rivers, WWTPs, and agriculture and aquaculture operation sites.
Soil resistome
Numerous studies have demonstrated that the natural soil environment is the origin of ARGs13,17,29. Many novel ARGs have been characterized from the soil environmental isolates or metagenomic sequences30,31,32. The presence of complex resistomes in permafrost and an isolated cave indicates that the environment is indeed the origin of ARGs12,14. The natural soil environment is regarded as a major reservoir of the antibiotic resistome, including intrinsic resistance and acquired resistances genes33; however, only some of these ARGs can acquire mobility and subsequently emerge in clinical settings, considering that mobile genetic elements (MGEs) syntenic with ARGs are rare in soil compared with pathogens18.
River resistome
Freshwater environments, including rivers, are considered reservoirs and dissemination routes for AMR34. Several studies have clearly depicted a larger increase in ARGs in human-impacted river sites than in the pristine river sites, indicating the effects of anthropogenic activities on the river resistome35,36,37. The river resistome is important for the potential reintroduction of AMR in humans because rivers are a major source of drinking water and irrigation water for agriculture38,39. Furthermore, in riverine ecosystems, the presence of novel ARG contexts, implying the evolution and adaptation of ARGs for transmission, can cause significant challenges when they are reintroduced into human populations40,41. The plausible factors influencing the river resistome have been analyzed in the contexts of the effects of pollution sources and the statistical evaluation of critical factors among various anthropogenic activities. Regarding pollution sources, the effects of WWTPs on the river resistome have been surveyed in various river systems because WWTPs are considered major pollution sources in rivers in urban regions42. In general, assessments of resistomes in upstream and downstream regions of particular river sites, to which WWTP effluents are exposed, revealed that effluents certainly affect the diversity and abundance of the river resistome43,44,45. For statistical elucidation, correlations between the river resistome and various factors were evaluated. Antibiotic residues, microbial community structures, environmental parameters, and socioeconomic factors related to anthropogenic activities were found to be associated with ARG increases and alterations in the diversity of the river resistome37,46,47. Moreover, fecal pollution has been identified as one of the major contributors to changes in the river resistome by tracking the genetic markers of human fecal contamination, such as crAssphage or fecal specific bacterial taxa37,48,49. However, a recent study reported that fecal contamination could not fully explain the dramatic ARG increase downstream of a heavily urbanized river, thereby indicating the proliferation of ARGs in the river environment via MGEs37. Unlike the natural soil resistome, the river resistome subjected to anthropogenic activity is not determined by the whole microbial community, indicating the prevalence of horizontal gene transfer (HGT) of ARGs among certain bacterial hosts18,37,50. Recent studies have also revealed a correlation between genetic traits and resistome changes, such as MGEs for HGT and metal resistance genes (MRGs) co-selected with ARGs, and the environmental hosts in which ARGs proliferate37,43,44,49,51,52. Among the MGEs, class 1 integrons have been demonstrated to be important molecular markers for resistome proliferation and anthropogenic pollution53,54. Some ARGs have been suggested as core resistome or critical ARGs responsible for altering the river resistome37. To prioritize critical factors affecting the river resistome, generalized approaches at the global scale and consolidation of the present river resistome data are required55.
Wastewater and WWTP
Wastewater and WWTPs are considered hotspots for the proliferation of antibiotic-resistant bacteria (ARB) and ARGs56. They are among the major interfaces between humans and the environment56 and are also recognized as hotspots for HGT of ARGs, resulting in the dissemination of AMR57. The presence of antibiotics, metals, and disinfectants in wastewater and WWTPs could act as selection pressures for ARG transfer and co-selection of ARGs with MRGs and disinfectant resistance genes, even though their concentrations are remarkably lower than therapeutic concentrations58. Recently, the mobile resistome in WWTPs comprised a major proportion of the WWTP resistome, resulting in the identification of critical ARGs in such environments59. The role of wastewater as a reservoir of ARGs is gaining substantial attention, considering the continuous reports on novel ARGs (some of which are already mobile)60; moreover, antibiotic-resistant pathogens and ARGs have recently emerged in the clinical setting61. The resistome in WWTPs according to the treatment steps has been intensively studied, and the persistence of ARGs throughout the treatment stages was elucidated62,63,64. A long-term investigation of WWTPs revealed remarkable changes in the resistome every 2–3 years, indicating successive selection of the resistome in activated sludge61. These efforts have focused on risk assessments of WWTPs; however, the consolidation of WWTP data from various regions and the establishment of standardized methodologies are required to derive the environmental framework for wastewater and WWTP resistomes56,57.
Agriculture
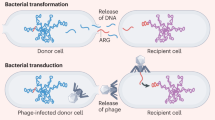
Previous studies have reported the importance of soil biota under the influence of anthropogenic activities in planetary health systems33. Organic manure and reclaimed wastewater have been identified as critical human and animal waste products that can induce alterations in the diversity and prevalence of ARGs and ARB in soil environments33,65. Agricultural sites (plant production environments) in which manure and wastewater are frequently used for soil amendment have been recognized as important routes for ARG transmission from humans and animals to the environment66. Moreover, introduced ARGs could be reintroduced into humans via agricultural products67. The FAO, WHO, and OIE emphasize the significance of the agricultural environment in ARG transmission (“Joint FAO/WHO expert meeting in collaboration with OIE on foodborne AMR: role of the environment, crops, and biocides”, 2019). Furthermore, recent studies have emphasized overall surveillance of ARGs in the agricultural environment and agricultural products to understand and control ARG transmission to humans68.
Aquaculture
Abundant use of antibiotics as prophylactic and therapeutic agents in aquaculture can induce selection, evolution, and HGT of ARB and ARGs in the environment69. Diverse microbiota, highly populated aquacultured animals, and the overuse of antibiotics designate aquaculture a “genetic hotspot” for gene exchange70. Several studies have reported an increase in ARGs in aquaculture systems71,72. The use of a specific antibiotic could promote ARGs against other classes of antibiotics, thereby indicating co-selection of ARGs mediated by MGEs72. Furthermore, the selected ARB and enriched ARGs in aquaculture could affect the microbiota and resistomes of adjacent environments73. The incidence of AMR in aquaculture can cause negative economic and social repercussions in these industries69. Enriched ARGs could be reintroduced into humans via adjacent environments and aquacultured animal products, suggesting an urgent need for surveillance of AMR.
Animal resistome
According to the WHO74, antibiotics are used in livestock animals for three purposes: therapeutic veterinary medicine, disease prevention (prophylaxis and metaphylaxis), and growth promotion. The latter two are strongly associated with the overuse of antibiotics in animals maintained under crowded conditions. For growth promotion, sublethal concentrations of antibiotics are widely used, although the mechanisms associated with weight gain remain unclear. Recent studies have reported that even low antibiotic concentrations affect resistome expansion in animals75. The use of antibiotics in livestock animals could trigger the selection of ARB and ARGs, which could subsequently be transferred to humans21.
The resistomes of various livestock animals have been investigated, and some regional studies have revealed a positive correlation between antibiotic use and resistome profiles as well as MGE dependency76. A recent study on the resistomes of slaughtered pigs and broilers across nine European countries revealed that the abundance and diversity of the fecal resistomes depended on antibiotic use across the nation77,78. In a subsequent and cross-sectional (human–animal) study, surveys of the resistomes of farm and slaughterhouse workers exposed to animals suggested that the human resistome was influenced by the animal resistome, thus, emphasizing the importance of surveillance for AMR in livestock animals79,80. Considering the complexity of the animal resistome and the presence of various external factors at livestock sites, systematic approaches should be implemented for further analysis.
In addition to livestock animals, wild animals are also known to be a source of ARG dissemination. The resistome of human-contacted wild animals revealed more-diverse ARGs than that of noncontacted animals81,82, thereby indicating that wild animals contribute to ARG transmission between humans and animals. Among wild animals, migratory birds were found to be responsible for ARG dissemination to the environment83. Moreover, the presence of pathogens carrying emerging ARGs in companion animals, such as dogs and cats, suggests that these animals act as ARB and ARG reservoirs84; however, resistome-level studies on these animals are scarce. Considering the proximity of companion animals to humans in daily life, the resistomes of these animals should be surveyed and assessed from the perspectives of the resistome and the One-Health to control ARG transmission to humans.
Human resistome
Metagenomics based on next-generation sequencing technology has been applied to the human microbiomes of the gut, skin, and respiratory tract to assess their resistomes. Understanding the dynamics of the human resistome and its relatedness to the other One-Health sectors is essential to control ARG flow from the other sectors to the human sector, particularly ARG transmission to disease-causing bacteria85. The resistome of the commensal bacterial community in the human microbiome is regarded as an important reservoir and dissemination route for clinical ARGs86. Several studies have elucidated the role of the human gut resistome by deciphering it and comparing its similarity with those of pathogens17,87,88. Although the transmission of ARGs from commensal bacteria to pathogens seems to be scarce89, the presence of almost identical ARGs and similar genetic contexts between the human gut and pathogenic bacteria indicate the importance of the human gut microbiome in the emergence of clinical ARGs. Correlations between the human gut resistome and the animal gut and environmental resistomes have been characterized79,90,91. Worldwide cohort studies revealed a nation-level resistome structure92,93, and antibiotic administration played a pivotal role in resistome and mobilome (a collection of all types of MGEs) structures94,95. Among the complex human gut resistomes, the importance of the mobile resistome, in which ARGs are associated with MGE in a genetic context, has been emphasized to understand ARG transmission from animals and the environment to pathogens88,96,97,98. Continuous identification of novel ARGs in the human gut proved the role of the human gut as an ARG reservoir and transmission route99. Furthermore, the establishment of the human gut resistome in infants, vertical transmission of the resistome and mobilome from mother to infant, and the correlations between the resistome and diseases have been studied to characterize the dynamics of the human gut resistome95,100,101,102.
The resistome of the human respiratory tract was assessed to understand the etiological cause and diagnosis of the AMR profile of infection103. In particular, resistance in the microbiome of the respiratory tract in patients with cystic fibrosis and chronic respiratory disease is of great concern due to frequent polymicrobial infection in these patients103. Moreover, respiratory tract infection in intensive care unit patients is a critical factor determining patient survival; however, current studies on the resistome of the respiratory tract are scarce, and further systematic approaches are required to characterize the resistome and provide guidance for proper antibiotic administration against such infections. The resistome of human skin of uncontacted Amerindian infants in a neonatal intensive care unit and hands exposed to the public metro system were characterized104,105,106, indicating a potential role of the skin microbiome in ARG transmission.
Future perspectives
Over the last decade, the paradigm of AMR research has changed, as the origin, transmission, and evolution of AMR have been discovered; however, as ARG transmission to humans from other One-Health sectors is only partially understood, it is expected that in the future, the following research approaches will be conducted based on the foundation of scientific achievements over the last decade. These attempts will improve our understanding of ARG transmission and guide strategies to mitigate AMR dissemination (Fig. 2).
Applications of cutting-edge technologies for antibiotic resistome studies
The antibiotic resistome has been recognized as dynamic and ever expanding11, thereby indicating the importance of cutting-edge technologies to elucidate its dynamics and to explore its diversity in depth. Newly developed next-generation sequencing methods and bioinformatics tools have accelerated the applications of such sequence-based methods for resistome studies in all One-Health sectors107. Metagenomics is an essential sequence-based approach to comprehend the complex resistome within the microbiome. Three different approaches, including functional metagenomics, shotgun metagenomics, and targeted gene sequencing (amplicon sequencing), have been applied in several studies, considering the trajectory of the studies over the last decade. Metagenomics has been widely used to identify novel ARGs (functional metagenomics)99,108 and ARG variants or genetic contexts of ARGs (targeted gene sequencing)31,41,109,110, and to analyze ARGs at the resistome level (shotgun metagenomics)37,111,112,113. According to specific research purposes, appropriate sequencing platforms, ARG databases, and analysis pipelines should be employed to understand AMR at the resistome level. A high-throughput quantitative polymerase chain reaction array, also termed the SmartChip system (Takara), has been widely used for resistome studies in various environmental microbiomes37,46,114,115. This system is advantageous, as a high number of ARGs can be analyzed simultaneously within a short time, and this system is more sensitive than the metagenomics approach for detecting ARGs; however, the limitation is that only previously characterized ARGs and MGEs can be detected, and information on the hosts carrying these genetic elements cannot be obtained. In the original primer version76, the primers were found to be redundant, and primers for newly discovered ARGs were absent. Recently, Primer set 2.0 was released to solve these issues116. Considering the cost, time, and sensitivity, this method can be widely used for resistome-level analysis if the continued development of primer sets is updated. For resistome analysis at the bacterial isolate level, whole-genome sequencing (WGS) for antibiotic susceptibility testing (AST) is gaining importance as a powerful approach to predict antibiotic susceptibility profiles at the genome level; this approach can be aided by machine learning and statistical models, although the inconsistency between the WGS method and culture-based AST needs to be improved117. In conclusion, these cutting-edge technologies will be essential for understanding ARG transmission between microbiomes or bacterial strains and will provide important information about the antibiotic resistome in the context of the One-Health approach.
Ranking the critical ARGs and their hosts
Since a complex resistome exists in all microbiomes, recent studies have attempted to classify particular ARGs that pose great threats to humans and other organisms. Their classification was revealed by understanding the core resistome, which is relatively stable in the microbiome, and the mobile resistome, which is genetically associated with MGEs37,88,118,119. Through these approaches, mobile ARGs in the microbiome have been recently reported119. Moreover, information about hosts of these mobile ARGs is essential to understand ARG carriers and their roles in transmission to humans37,96,113.
ARG transmission at the interfaces among One-Health sectors
It is important to understand ARG flow between the One-Health sectors by monitoring the interfaces among these sectors120. Thus, appropriate configuration of the associated testbeds and their interfaces is essential. If both culture-dependent and culture-independent methods are simultaneously employed in an appropriate manner to understand the resistome at the interfaces, then snapshots of transmission among the One-Health sectors can be obtained. Valuable information integrated into and accumulated through these approaches will reveal the actual dynamics of ARG transmission.
Selective pressures affecting the emergence, transmission, and evolution of ARGs
Recent studies have reported that anthropogenic activities shape the environmental resistome37,46,93; however, limited information about the specific factors that induce changes in the resistomes of One-Health sectors is available. To reveal the selective pressures causing such changes, it is necessary to collect relevant data in a more granular manner, and to compare the data with the resistome data in a reliable statistical way. Furthermore, monitoring changes in the resistome in response to individual selective pressure in microcosm or mesocosm studies are required to obtain experimental evidence. A systematic understanding of the correlation between selective pressure and resistome changes resulting from mitigation will be an essential prerequisite for mitigating ARG transmission to humans.
Mechanisms overcoming taxonomic barriers in ARG transmission
AMR has long been present in antibiotic-producing bacteria, typically nonpathogenic environmental bacteria6,7,121; however, at some point, after crossing over into pathogens or related strains through evolutionary events that crossed taxonomic barriers, ARGs became mobile and emerged in the clinical setting121. No clear mechanism has been clearly identified yet, although a case study suggested a “carry-back” mechanism for ARG transfer from antibiotic producers to pathogens122. Considering the substantial number of environmental ARGs that are not yet mobile, the mechanisms of taxonomic barrier crossing in ARG transmission need to be better understood.
Conclusion
The concept of the antibiotic resistome and a One-Health approach is crucial to understand and mitigate ARG transmission between the One-Health sectors. Although important findings of AMR in these sectors have been recently unveiled, various factors need to be explored to understand the origin, emergence, dissemination, and evolution of ARGs. Based on cutting-edge technologies, several research topics suggested in the present study need to be further explored in the near future. Such endeavors will facilitate our hopes to overcome the arms race between antibiotics and AMR.
References
Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull. World Health Organ. 79, 780–790 (2001).
Yong, D. et al. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054 (2009).
Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 (2016).
Rammelkamp, C. H. & Maxon, T. Resistance of Staphylococcus aureus to the Action of Penicillin. Proc. Soc. Exp. Biol. Med. 51, 386–389 (1942).
Stekel, D. First report of antimicrobial resistance pre-dates penicillin. Nature 562, 192 (2018).
Benveniste, R. & Davies, J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 70, 2276–2280 (1973).
D’Costa, V. M., McGrann, K. M., Hughes, D. W. & Wright, G. D. Sampling the antibiotic resistome. Science 311, 374–377 (2006).
Wright, G. D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 (2007).
Wright, G. D. Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594 (2010).
Wright, G. D. The antibiotic resistome. Expert Opin. Drug Discov. 5, 779–788 (2010).
Perry, J. A., Westman, E. L. & Wright, G. D. The antibiotic resistome: what’s new? Curr. Opin. Microbiol. 21, 45–50 (2014).
D’Costa, V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011).
Finley, R. L. et al. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710 (2013).
Pawlowski, A. C. et al. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 7, 13803 (2016).
D’Costa, V. M., Griffiths, E. & Wright, G. D. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489 (2007).
Dantas, G. & Sommer, M. O. Context matters - the complex interplay between resistome genotypes and resistance phenotypes. Curr. Opin. Microbiol. 15, 577–582 (2012).
Forsberg, K. J. et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111 (2012).
Forsberg, K. J. et al. Bacterial phylogeny structures soil resistomes across habitats. Nature 509, 612–616 (2014).
Perry, J. A. & Wright, G. D. Forces shaping the antibiotic resistome. Bioessays 36, 1179–1184 (2014).
Perry, J. A. & Wright, G. D. The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front. Microbiol. 4, 138 (2013).
Hu, Y., Gao, G. F. & Zhu, B. The antibiotic resistome: gene flow in environments, animals and human beings. Front. Med. 11, 161–168 (2017).
Rubin, C. et al. Review of institute of medicine and national research council recommendations for one health initiative. Emerg. Infect. Dis. 19, 1913–1917 (2013).
Gronvall, G., Boddie, C., Knutsson, R. & Colby, M. One health security: an important component of the global health security agenda. Biosecur. Bioterror. 12, 221–224 (2014).
World Health Organization, Food, Agriculture Organization of the United Nations & World Organisation for Animal Heallth. High-level technical meeting to address health risks at the human-animal ecosystems interfaces. Mexico city, Mexico 15–17 November 2011.
Kahn, L. H. Antimicrobial resistance: a One Health perspective. Transs R. Soc. Trop. Med. Hyg. 111, 255–260 (2017).
Van Puyvelde, S., Deborggraeve, S. & Jacobs, J. Why the antibiotic resistance crisis requires a One Health approach. Lancet Infect. Dis. 18, 132–134 (2018).
Hernando-Amado, S., Coque, T. M., Baquero, F. & Martínez, J. L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442 (2019).
Martinez, J. L. Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367 (2008).
Riesenfeld, C. S., Goodman, R. M. & Handelsman, J. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6, 981–989 (2004).
Spanogiannopoulos, P., Thaker, M., Koteva, K., Waglechner, N. & Wright, G. D. Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes. Antimicrob. Agents Chemother. 56, 5061–5069 (2012).
Kim, D. W., Thawng, C. N., Choi, J. H., Lee, K. & Cha, C. J. Polymorphism of antibiotic-inactivating enzyme driven by ecology expands the environmental resistome. ISME J. 12, 267–276 (2018).
Berglund, F. et al. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome 7, 52 (2019).
Zhu, Y. G. et al. Soil biota, antimicrobial resistance and planetary health. Environ. Int. 131, 105059 (2019).
Nnadozie, C. F. & Odume, O. N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 254, 113067 (2019).
Chen, B. et al. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ. Sci. Technol. 47, 12753–12760 (2013).
Ouyang, W. Y., Huang, F. Y., Zhao, Y., Li, H. & Su, J. Q. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 99, 5697–5707 (2015).
Lee, K. et al. Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome 8, 2 (2020).
Han, Z. et al. Antibiotic resistomes in drinking water sources across a large geographical scale: Multiple drivers and co-occurrence with opportunistic bacterial pathogens. Water Res. 183, 116088 (2020).
Gudda, F. O. et al. Antibiotic-contaminated wastewater irrigated vegetables pose resistance selection risks to the gut microbiome. Environ. Pollut. 264, 114752 (2020).
Amos, G. C. A. et al. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J. 12, 681–691 (2018).
Böhm, M. E., Razavi, M., Marathe, N. P., Flach, C. F. & Larsson, D. G. J. Discovery of a novel integron-borne aminoglycoside resistance gene present in clinical pathogens by screening environmental bacterial communities. Microbiome 8, 41 (2020).
Aristi, I. et al. Mixed effects of effluents from a wastewater treatment plant on river ecosystem metabolism: subsidy or stress? Freshw. Biol. 60, 1398–1410 (2015).
Marathe, N. P. et al. Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Res. 124, 388–397 (2017).
Lekunberri, I., Balcázar, J. L. & Borrego, C. M. Metagenomic exploration reveals a marked change in the river resistome and mobilome after treated wastewater discharges. Environ. Pollut. 234, 538–542 (2018).
Wang, J. Y., An, X. L., Huang, F. Y. & Su, J. Q. Antibiotic resistome in a landfill leachate treatment plant and effluent-receiving river. Chemosphere 242, 125207 (2020).
Zhu, Y. G. et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2, 16270 (2017).
Milaković, M. et al. Pollution from azithromycin-manufacturing promotes macrolide-resistance gene propagation and induces spatial and seasonal bacterial community shifts in receiving river sediments. Environ. Int. 123, 501–511 (2019).
Karkman, A., Pärnänen, K. & Larsson, D. G. J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 10, 80 (2019).
Chen, H. et al. Characterization and source-tracking of antibiotic resistomes in the sediments of a peri-urban river. Sci. Total Environ. 679, 88–96 (2019).
Peng, F. et al. Urbanization drives riverine bacterial antibiotic resistome more than taxonomic community at watershed scale. Environ. Int. 137, 105524 (2020).
Bai, Y., Ruan, X., Xie, X. & Yan, Z. Antibiotic resistome profile based on metagenomics in raw surface drinking water source and the influence of environmental factor: a case study in Huaihe River Basin, China. Environ. Pollut. 248, 438–447 (2019).
Chen, H., Chen, R., Jing, L., Bai, X. & Teng, Y. A metagenomic analysis framework for characterization of antibiotic resistomes in river environment: application to an urban river in Beijing. Environ. Pollut. 245, 398–407 (2019).
Amos, G. C. et al. Validated predictive modelling of the environmental resistome. ISME J. 9, 1467–1476 (2015).
Gillings, M. R. et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279 (2015).
Stedtfeld, R. D. et al. Antimicrobial resistance dashboard application for mapping environmental occurrence and resistant pathogens. FEMS Microbiol. Ecol. 92, fiw020 (2016).
Karkman, A., Do, T. T., Walsh, F. & Virta, M. P. J. Antibiotic-rresistance genes in waste water. Trends Microbiol. 26, 220–228 (2018).
Berendonk, T. U. et al. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13, 310–317 (2015).
Andersson, D. I. & Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478 (2014).
Che, Y. et al. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 7, 44 (2019).
Marathe, N. P. et al. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7, 97 (2019).
Yin, X. et al. Exploration of the antibiotic resistome in a wastewater treatment plant by a nine-year longitudinal metagenomic study. Environ. Int. 133, 105270 (2019).
Huyan, J. et al. Dynamics of class 1 integrons in aerobic biofilm reactors spiked with antibiotics. Environ. Int. 140, 105816 (2020).
Yoo, K., Yoo, H., Lee, J., Choi, E. J. & Park, J. Exploring the antibiotic resistome in activated sludge and anaerobic digestion sludge in an urban wastewater treatment plant via metagenomic analysis. J. Microbiol. 58, 123–130 (2020).
Ng, C. et al. Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front. Microbiol. 10, 172 (2019).
Udikovic-Kolic, N., Wichmann, F., Broderick, N. A. & Handelsman, J. Bloom of resident antibiotic-resistant bacteria in soil following manure fertilization. Proc. Natl. Acad. Sci. USA 111, 15202–15207 (2014).
Chen, C. et al. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ. Int. 128, 233–243 (2019).
Blau, K. et al. The transferable resistome of produce. mBio 9, e01300–e01318 (2018).
Wang, Y. et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2, 16260 (2017).
Cabello, F. C., Godfrey, H. P., Buschmann, A. H. & Dölz, H. J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 16, e127–e133 (2016).
Watts, J. E. M., Schreier, H. J., Lanska, L. & Hale, M. S. The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Mar. Drugs 15, 158 (2017).
Fang, H. et al. Metagenomic analysis of bacterial communities and antibiotic resistance genes in the Eriocheir sinensis freshwater aquaculture environment. Chemosphere 224, 202–211 (2019).
Zeng, Q., Liao, C., Terhune, J. & Wang, L. Impacts of florfenicol on the microbiota landscape and resistome as revealed by metagenomic analysis. Microbiome 7, 155 (2019).
Muziasari, W. I. et al. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 92, fiw052 (2016).
World Health Organization. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals (2017).
You, Y. & Silbergeld, E. K. Learning from agriculture: understanding low-dose antimicrobials as drivers of resistome expansion. Front. Microbiol. 5, 284 (2014).
Looft, T. et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 109, 1691–1696 (2012).
Munk, P. et al. Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat. Microbiol. 3, 898–908 (2018).
Luiken, R. E. C. et al. Associations between antimicrobial use and the faecal resistome on broiler farms from nine European countries. J. Antimicrob. Chemother. 74, 2596–2604 (2019).
Van Gompel, L. et al. Description and determinants of the faecal resistome and microbiome of farmers and slaughterhouse workers: a metagenome-wide cross-sectional study. Environ. Int. 143, 105939 (2020).
Van Gompel, L. et al. Occupational exposure and carriage of antimicrobial resistance genes (tetW, ermB) in pig slaughterhouse workers. Ann. Work Expo. Health 64, 125–137 (2020).
Allen, H. K. et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259 (2010).
Tsukayama, P. et al. Characterization of wild and captive baboon gut microbiota and their antibiotic resistomes. mSystems 3, e00016–e00018 (2018).
Cao, J. et al. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 8, 26 (2020).
Pomba, C. et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 72, 957–968 (2017).
Wright, G. D. Environmental and clinical antibiotic resistomes, same only different. Curr. Opin. Microbiol. 51, 57–63 (2019).
Surette, M. D. & Wright, G. D. Lessons from the environmental antibiotic resistome. Annu. Rev. Microbiol. 71, 309–329 (2017).
Sommer, M. O. A., Dantas, G. & Church, G. M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131 (2009).
Raymond, F. et al. Culture-enriched human gut microbiomes reveal core and accessory resistance genes. Microbiome 7, 56 (2019).
van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140087 (2015).
Sun, J. et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 11, 1427 (2020).
Pehrsson, E. C. et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature 533, 212–216 (2016).
Hu, Y. et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 4, 2151 (2013).
Forslund, K., Sunagawa, S., Coelho, L. P. & Bork, P. Metagenomic insights into the human gut resistome and the forces that shape it. Bioessays 36, 316–329 (2014).
Willmann, M. et al. Distinct impact of antibiotics on the gut microbiome and resistome: a longitudinal multicenter cohort study. BMC Biol. 17, 76 (2019).
Gasparrini, A. J. et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 4, 2285–2297 (2019).
Bertrand, D. et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat. Biotechnol. 37, 937–944 (2019).
Hu, Y. et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl. Environ. Microbiol. 82, 6672–6681 (2016).
Ravi, A. et al. The commensal infant gut meta-mobilome as a potential reservoir for persistent multidrug resistance integrons. Sci. Rep. 5, 15317 (2015).
Moore, A. M. et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS ONE 8, e78822 (2013).
Pärnänen, K. et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 9, 3891 (2018).
Loo, E. X. L. et al. Longitudinal assessment of antibiotic resistance gene profiles in gut microbiomes of infants at risk of eczema. BMC Infect. Dis. 20, 312 (2020).
Gibson, M. K., Crofts, T. S. & Dantas, G. Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 27, 51–56 (2015).
Sherrard, L. J., Tunney, M. M. & Elborn, J. S. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 384, 703–713 (2014).
Clemente, J. C. et al. The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015).
Hourigan, S. K. et al. Comparison of infant gut and skin microbiota, resistome and virulome between neonatal intensive care unit (NICU) environments. Front. Microbiol. 9, 1361 (2018).
Kang, K. et al. The environmental exposures and inner- and intercity traffic flows of the metro system may contribute to the skin microbiome and resistome. Cell Rep. 24, 1190–1202 (2018).
Crofts, T. S., Gasparrini, A. J. & Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434 (2017).
Pehrsson, E. C., Forsberg, K. J., Gibson, M. K., Ahmadi, S. & Dantas, G. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front. Microbiol. 4, 145 (2013).
Böhm, M. E., Razavi, M., Flach, C. F. & Larsson, D. G. J. A novel, integron-regulated, class C β-lactamase. Antibiotics(Basel) 9, 123 (2020).
Ma, L., Li, A. D., Yin, X. L. & Zhang, T. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ. Sci. Technol. 51, 5721–5728 (2017).
Port, J. A., Wallace, J. C., Griffith, W. C. & Faustman, E. M. Metagenomic profiling of microbial composition and antibiotic resistance determinants in Puget Sound. PLoS ONE 7, e48000 (2012).
Kristiansson, E. et al. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE 6, e17038 (2011).
Ma, L. et al. Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken, and human feces. Environ. Sci. Technol. 50, 420–427 (2016).
Wang, F. H. et al. High throughput profiling of antibiotic resistance genes in urban park soils with reclaimed water irrigation. Environ. Sci. Technol. 48, 9079–9085 (2014).
Chen, Q. L. et al. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 51, 8149–8157 (2017).
Stedtfeld, R. D. et al. Primer set 2.0 for highly parallel qPCR array targeting antibiotic resistance genes and mobile genetic elements. FEMS Microbiol. Ecol. 94, fiy130 (2018).
Schürch, A. C. & van Schaik, W. Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance. Ann. N. Y. Acad. Sci. 1388, 108–120 (2017).
Munck, C. et al. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 6, 8452 (2015).
Zhang, A. N., Hou, C. J., Negi, M., Li, L. G. & Zhang, T. Online searching platform for the antibiotic resistome in bacterial tree of life and global habitats. FEMS Microbiol Ecol. 96, fiaa107 (2020).
Ma, Z., Lee, S. & Jeong, K. C. Mitigating antibiotic resistance at the livestock-environment interface: a review. J. Microbiol. Biotechnol. 29, 1683–1692 (2019).
Martinez, J. L., Coque, T. M. & Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123 (2015).
Jiang, X. et al. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8, 15784 (2017).
Acknowledgements
This work was supported by the Korea Ministry of Environment (MOE) as “the Environmental Health Action Program (2016001350004)” and by the Korea Centers for Disease Control and Prevention (2017ER540701).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, DW., Cha, CJ. Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. Exp Mol Med 53, 301–309 (2021). https://doi.org/10.1038/s12276-021-00569-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-021-00569-z
This article is cited by
-
Genomic surveillance for antimicrobial resistance — a One Health perspective
Nature Reviews Genetics (2024)
-
Effects of a Mining Dam Disaster on Antimicrobial-Resistant and Metal-Tolerant Bacterial Strains Recovered from Environmental Samples
Water, Air, & Soil Pollution (2024)
-
“One Health” perspective on prevalence of co-existing extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae: a comprehensive systematic review and meta-analysis
Annals of Clinical Microbiology and Antimicrobials (2023)
-
Lactiplantibacillus plantarum, lactiplantibacillus pentosus and inulin meal inclusion boost the metagenomic function of broiler chickens
Animal Microbiome (2023)
-
Latent antibiotic resistance genes are abundant, diverse, and mobile in human, animal, and environmental microbiomes
Microbiome (2023)