Abstract
Five decades of research and practical application of computers in biomedicine has given rise to the discipline of medical informatics, which has made many advances in genomic and translational medicine possible. Developments in nanotechnology are opening up the prospects for nanomedicine and regenerative medicine where informatics and DNA computing can become the catalysts enabling health care applications at sub-molecular or atomic scales. Although nanomedicine promises a new exciting frontier for clinical practice and biomedical research, issues involving cost-effectiveness studies, clinical trials and toxicity assays, drug delivery methods, and the implementation of new personalized therapies still remain challenging. Nanoinformatics can accelerate the introduction of nano-related research and applications into clinical practice, leading to an area that could be called “translational nanoinformatics.” At the same time, DNA and RNA computing presents an entirely novel paradigm for computation. Nanoinformatics and DNA-based computing are together likely to completely change the way we model and process information in biomedicine and impact the emerging field of nanomedicine most strongly. In this article, we review work in nanoinformatics and DNA (and RNA)-based computing, including applications in nanopediatrics. We analyze their scientific foundations, current research and projects, envisioned applications and potential problems that might arise from them.
Similar content being viewed by others
Main
Over the past two decades, bioinformatics and systems biology have addressed informational challenges in biomedicine at the molecular and cellular level, leading to the sequencing of the Human Genome and other -omics projects. At a higher scale, or organismal level, medical informatics deals with patient information, whereas public health informatics focuses on their aggregation at the population level. Recently, Biomedical Informatics (BMI) has emerged to integrate the fields of Medical Informatics and Bioinformatics. However, new informatics methods will be needed to deal with phenomena and research at the smallest, submolecular or atomic levels. The new discipline of Nanoinformatics aims to represent and work with information at the nano level. In this review article, we use the generic name, “Nanoinformatics,” to avoid any premature overspecialization in a discipline—like medical nanoinformatics or bionanoinformatics—that still needs to be more completely defined. We include in this review the related computational field dealing with methods and devices that process biological information by using DNA and RNA.
Nanoinformatics refers to the use of informatics techniques for analyzing and processing information about the structure and physico-chemical characteristics of nanoparticles, their environments, and applications. Figure 1 schematically represents some of its relationships with related disciplines.
Nanoinformatics is a newly emerging informatics area, working at the intersection between informatics (computer science and information technologies), nanotechnology, medicine and established areas such as biology, chemistry, and physics. The first large official meeting in the field was held in Virginia in 2007, with support from the US National Science Foundation (NSF) (http://128.119.56.118/~nnn01/Workshop.html). In 2008, the European Commission launched the first European initiative linking Nanoinformatics and BMI, the ACTION Grid project in which the authors participate (http://www.action-grid.eu/). These and other foundational initiatives have emerged to support Nanoinformatics as a discipline designed to catalyze and accelerate research and developments in nanomedicine (Baker NA, Fritts M, Guccione S, Paik DS, Pappu RV, Patri A, Rubin D, Shaw SY, Thomas DG 2009 “Nanotechnology Informatics”, White paper commissioned by the US National Cancer Institute, NCI, Bethesda, MD).
Nanotechnology in Medicine: Physical Foundations of Nanomedicine
A search in Medline for the term “nanomedicine” at the time of writing shows 1224 articles already published. Some examples are highlighted in Table 1, drawn from a wide range of sources (1–11).
Despite the numerous current applications, the biological effects of these nano-scale devices are far from completely understood, and research in this field is still in its infancy. The physico-chemical properties of nanoparticles such as their volume, shape, 3-D configuration, flexibility, electrostatic properties and purity, among others, can strongly influence their biological interactions in different system, tissue, organ, cell, organelle, and molecular environments. A nanoparticle can therefore be designed to achieve specific medical goals based on its novel nanoscale properties coupled with a functionalization that facilitates its interaction with the different biological environments it will encounter. This sensitivity of the efficacy of a nanoparticle formulation to its detailed structure is both its advantage and its handicap, because nanoparticles are generally polydisperse in structure: there may be large lot-to-lot variability and the subpopulation structures may induce different effects. Insufficient characterization and control of the structure of nanomaterials have been and continue to be major problems in the development and testing of nanoparticle formulations. This suggests profound medical implications for nanoparticles and nanodevices, which will require understanding critical issues such as the potential toxic or therapeutic effect of nanoparticles. To effectively and efficiently manage all these data, new informatics tools must be implemented to archive, access, and annotate the relevant data, to couple that data to new computational methods, and to model the relation between nanoparticle structures and their effects in biological environments, so that their cumulative effect in tissue can be established and linked eventually to structure/activity relationships and clinical outcomes.
Some nanoparticles and nanodevices have been already approved by the Food and Drug Administration or are in advanced clinical trials. For instance, superparamagnetic nanoparticles to detect metastatic lymph node involvement in a variety of solid tumors (12); albumin-nanoparticles bearing paclitaxel for metastatic breast cancer (13); or new devices combining microfluidics and nanosensors are being investigated for the detection of circulating tumor cells and biomarkers from peripheral blood (14,15).
These applications of nanomaterials offer new challenges for personalized medicine. Personalized medicine aims to adapt therapies to individual patients (16). Thus, drugs may be tailored to groups of people with similar or related genetic or physiologic characteristics. In this context, classical clinical studies must be redesigned to adapt to the advances made in genomics, proteomics, and pharmacogenetics. The introduction of nanoparticles that can target different molecules or groups of atoms with high precision can significantly advance personalization of clinical procedures. Examples include elucidating a molecular phenotype or fingerprint of a patient's cancer, delivering molecular targeted therapies, monitoring the efficacy of treatment, and informing future treatment decisions (R Cachau and M Fritts, unpublished data, 2009). The specialty of pediatrics involves unique developmental issues, which will require considerable targeted research, because
-
Childhood metabolism is different, with specific excretion profiles, and use of different receptors.
-
The patient population is smaller and with special characteristics, which affects the design of clinical trials and may reduce potential markets for drugs.
-
The immune system works differently in children.
-
Diagnosis and treatment are more difficult to implement.
-
Nanoparticles as contrast agents for diagnostic imaging may have different effects in children than in adults.
-
Nanomaterials need to be designed for lower toxicities and greater effectiveness in children.
A number of initiatives already apply nanomedicine to pediatrics (17). For example, Mattel Children's Hospital at UCLA has launched a pioneer NanoPediatrics Program, attempting to personalize medicine for children (http://www.nanopediatrics.ucla.edu/). At the same institution, other groups are trying to improve the delivery of anti-cancer agents to tumor cells in children, so as to increase the effectiveness of chemotherapy and reduce its toxicity.
The company NanoStar Health Corporation is developing novel nanotechnology methods for instantly assessing the metabolic profile of premature infants without periodically collecting a blood sample from a newborn's heel (http://www.nanostarhealth.com/Welcome.html).
The company Genetic Immunity reported its DermaVir Patch, a Nanomedicine Vaccine product candidate for the treatment of HIV/AIDS in children (http://www.geneticimmunity.com/GI00.html).
A new generation of nanodevices for diagnosing and treating children with genetic diseases and cancer includes the use of nanoparticles for diagnostic imaging during pregnancy, nano-based newborn screening tests for genetic abnormalities and mutation detection for cystic fibrosis using a nanoparticle based bio-sensing system (18). Similarly, nanomechanical approaches study the effect of drugs on Pseudomonas aeruginosa, the causative agent of chronic lung infections in patients affected by cystic fibrosis (19).
Nanoinformatics can contribute critically to some of the above. Examples include: describing the use of computer simulations for improving targeted delivery of magnetic aerosol droplets to specific lung regions to treat asthma, cystic fibrosis, respiratory infection, or lung cancer (20). Stone et al. analyzed the potential toxicity of air pollution nanoparticles in children and adults in terms of cellular and molecular interactions involved in inducing oxidative stress and inflammation (21,22).
Managing Toxicity of Nanomaterials
The scientific community faces potentially serious issues related to patient safety and possible secondary effects related to the use of nanoparticles. Nanoparticles can have a natural or biological origin—like viruses—or be engineered, including very different elements. It is their small size that gives nanoparticles their unique electrical, optical, and chemical properties, raising concerns about their potential toxicity (23).
More research on the physico-chemical properties of nanomaterials, such as size, shape, crystalline structure, chemical composition, and how all these present, whether in vivo or in vitro, are necessary. In this regard, controversies have arisen when research articles suggest that nanoparticles can damage DNA or specific cells. In some cases, studies in vitro may have little relevance to human exposure risks or may be even deeply flawed. In some cases, it is the dose and mechanism of action that makes a nanoparticle therapy toxic, rather than the properties of the nanoparticle itself.
In Nanoinformatics, toxicity will play a critical role. There are already databases of toxic effects such as National Institute for Occupational Safety and Health's (NIOSH) (http://www.cdc.gov/niosh/), and Oregon Nanoscience and Microtechnologies Institute's (ONAMI) toxicity screen using embryonic zebra fish (http://www.greennano.org). Researchers will use nanoinformatics to model and simulate toxicity processes, linking them with actual patient data from computerized medical records to predict human response. In Europe and the United States some initiatives—like Advancing Clinico Genomic Trials on Cancer (ACGT) (24) and the cancer Biomedical Informatics Grid (caBIG) (25)—have developed new approaches to data sharing, modeling, and simulation of drug delivery and distributed computing processing over sophisticated Grid-based infrastructures. They aim to accelerate the trials needed to ensure drug efficacy and safety. By using advanced computational methods, researchers can reduce the time taken to translate drugs—and nanoparticles—from the laboratory to clinical practice.
Nanoinformatics: Informational Foundations for Nanomedicine
Over the past few years, the field of data integration has evolved from its original clinical and genomic emphases to address multilevel integration down to the molecular level. For instance, we have performed research to integrate genomic with medical information systems (26,27). This process has proven to be more difficult than anticipated, because of a variety of reasons.
From a computational perspective, data integration at the nano level poses even more difficult informational challenges that must be addressed—parallel to those faced at higher scales by bioinformatics and medical informatics. These include, for instance:
-
The development of central repositories of nanoparticle data [such as GenBank for nucleotide sequences or Protein Data Bank (PDB) for structures], and specific databases (the thousand plus public databases with –omics data).
-
Standards for information storage and exchange at the nano level (such as HL7 for health information or DICOM for images).
-
Domain nano-ontologies—like Gene Ontology for genomics—terminologies, and vocabularies—like Logical Observation Identifiers Names and Codes (LOINC) for laboratory data, Systematized Nomenclature of Medicine (SNOMED) for clinical terminology or the Unified Medical Language System.
-
Tools for decision support—like expert systems or those based on data mining but working on an even more rapidly changing knowledge-base of nanomedicine.
For nanomedicine research, there are today many disparate sources of information. For instance, researchers at the University of Talca, in collaboration with members of the Advanced Biomedical Computing Center at the NCI, have developed a pilot database of nanoparticle structures, currently containing 5 different categories: carbon nanotubes, dendrimers, buckyballs, gold particles, and mag probes. To include a new nanoparticle structure, 3 steps are followed: 1) insert a new molecular structure for the nanoparticle, 2) simulate the structure in a specific realistic biological environment, and 3) display time dependent 3-D renderings of the nanoparticles during simulation, and extract useful data from the simulations and the nanoparticle conformations and reactions with the environment to aid in extracting candidate structural motifs responsible for their particular activity. The database contains 4 categories of data: properties (authors, physical and chemical properties), low-energy configurations, associated files, and technical information. Researchers can contribute by adding new nanoparticle structures and information to the database, by using available structures in additional computations, in annotating existing structures and results, and by initiating collaborative modeling exercises through the use of a wiki interface (http://nanobiology.utalca.cl/csn/index2.php).
A different approach to building a classification of nanoparticles based on nanoparticle constituent structures and their properties rather than structural motifs has been proposed by Tomalia (28). On the basis of intrinsic properties of nanoelements and nanocompounds, Tomalia anticipates a future nanoperiodic table (or tables). The European Commission and several US agencies have launched a collaboration for the development of inventories of nanoparticles, including techniques for modeling relationships between nanoparticle properties and toxicity and the interaction of nanoparticles with biological systems (http://cordis.europa.eu/nanotechnology/src/safety.htm). These and other initiatives will allow, for instance, to eventually link toxicological effects to patient data from computerized medical records. Therefore, it can be expected that a large number of information resources will be soon available for researchers.
To locate information that is available at multiple sites over the Web, bioinformaticians have developed catalogues and inventories of resources, such as BioPortal (29), the Bioinformatics Links Directory (BLD) (30) or the catalogue available at the European Bioinformatics Institute (http://www.ebi.ac.uk/services/). Based on text mining techniques, the authors have built a new approach to automatically create a searchable index of bioinformatics resources, with information automatically extracted from abstracts of research articles using text mining techniques (31). We are currently building a nanoinventory of resources from the literature (32), in which we have seen the great variability in how researchers report their results. In this regard, the bioinformatics community has worked to develop consensus towards publishing-omics information in the literature, so studies, design descriptions, methods, results, and structured abstracts follow standard procedures for publication. This has yet to be done for Nanoinformatics (33). Table 2 presents some examples of current resources in the nano areas.
Because nanotechnology involves a lengthy development pipeline from nanomaterial synthesis to physical characterization, to in vitro and in vivo characterization, and to clinical application, information integration will be key to addressing the needs of this community. Beyond that, dry bench, or in silico methods, will begin to complement wet bench research, although an enormous amount of heterogeneous empirical data must be collected to produce results from these methods. This will be one of the central challenges for nanoinformatics.
Previous work in the -omics area have shown that data cannot be automatically integrated. For instance, with microarray area, minimum information standards [e.g., the minimum information about a microarray experiment (MIAME) that is required to enable the interpretation of the results of the microarray experiments] specify basic procedures for collecting and sharing data from diverse laboratories (34). In 2008, the Minimum Information for Nanomaterial Characterization (MINChar) Initiative launched a process for developing a similar standard for nanotoxicology studies (http://characterizationmatters.org/).
From a computational perspective, heterogeneity must be addressed at both the syntactic (hardware and software) and semantic levels. For instance, some medical information systems can run on PCs, with windows, whereas others run on UNIX workstations. Furthermore, semantic differences can prevent automatic integration. For instance, what is named “fever” in one database can be named “febrile process” in another one. The Unified Medical Language System has been developed to be a common vocabulary framework to help computers communicate and interoperate. Ontologies, which describe classes—e.g., species, organisms, and their organs, tissues, and cells—with their properties and relationships in real-world contexts, have proven to be an important computational approach for systematizing knowledge—particularly in biomedicine (35). They have become a fundamental technology for structuring knowledge and, therefore, facilitating system interoperability, information retrieval and more semantically informed search. Thus, they are central to the development of the Semantic Web (http://www.w3.org/).
Structuring information in nanomedicine is essential for advancing research, and for this, controlled vocabularies and ontologies will be key, but present challenging definitional and representational problems. The MeSH term “nanoparticle” was introduced in 2007 (http://www.ncbi.nlm.nih.gov/sites/entrez). In addition, new taxonomies and ontologies such as the Nanomedicine Taxonomy (NT) (Gordon, N., Sagman, U. 2003 Nanomedicine Taxonomy. Canadian Institute of Health Research & Canadian NanoBusiness Alliance, Toronto, Canada) and the NanoParticle Ontology (NPO) are being developed (36). NT is centered around a structured description of the areas included within nanomedicine. NPO is an ontology designed to capture knowledge related to nanoparticles, based on their physical, chemical, and biological/functional characterization. NPO is being developed at Washington University and has already been adopted in different initiatives of the NCI. Because NPO is a domain ontology, there are proposals to develop domain ontologies and controlled vocabularies in the area of cancer nanotechnology, with support from the NCI, and other initiatives, such as an ontology for discovery of new nanomaterials, a functional ontology, the Nanotech Index Ontology, an atlas of nanotechnology or BiomedGT, to translate among different ontologies (NCI 2009 white paper, previously mentioned).
Ontologies are also important for developing new Nanoportals, after other successful BMI initiatives such as BIOPORTAL (29). These kinds of nanoportals will benefit from recent research on standards for interoperability, text mining, and collaborative approaches such as those proposed in the framework of the social-network oriented Web 2.0. In this context, distributed researchers can collaborate remotely to develop ontologies, databases, modeling, and simulation software tools or even engineer nanoparticles. All this information can be shared, reused, and discussed and analyzed by other users later, leading to new scientific insights (37). Figure 2 shows a representation of the infrastructure and computational tasks required in the nano areas.
An example is the cancer Nanotechnology Laboratory portal (caNanoLab) (https://wiki.nci.nih.gov/display/ICR/caNanoLab). It provides a secure support for data curation, annotating nanoparticles with characterizations derived from physical and in vitro nanoparticle assays and the sharing of these characterizations and related protocols. caNanoLab has developed through collaboration between the NCI Center for Biomedical Informatics and Information Technology (CBIIT), which developed caBIG and NCI's Alliance for Nanotechnology in Cancer (http://nano.cancer.gov), which includes the Cancer Centers of Nanotechnology Excellence (CCNEs) and the Nanotechnology Characterization Laboratory (NCL). The Collaboratory for Structural Nanobiology (CSN) is a pilot, which supports sharing of structural models of nanoparticles and predictive computational modeling.
Translational Nanoinformatics: New Challenges
As noted above, many current nanoinformatics applications look very similar—at least, on the surface—to comparable systems already built in medical and bioinformatics. In this sense, one can identify a continuum of BMI approaches from macro down to the nano dimensions. But, physical laws and the emerging properties of biological components are different at the different levels, from atoms to populations, bringing unique challenges to informatics. This suggests that new approaches are needed for data and knowledge integration at the nano level.
A broad, integrative vision of BMI implies a strong need to link and integrate molecular and clinical data for scientific discovery. Given that many molecular level processes increasingly involve new atomic-level or nano-level measurements and understanding, translational bioinformatics aims to translate the increasing amount of -omics data into new knowledge than can provide, among other results, personalized medical diagnosis and therapy for patients, despite the complexity of multi-gene and environmental interactions in human diseases. From such a viewpoint, nanomedicine requires novel insights beyond the current technology of informatics typically focused on collecting, representing and linking information, and managing various aspects of both system and semantic heterogeneity.
In the nanoscience field, when scientists face problems at the 10 ångströms scale, unexpected scientific phenomena emerge, involving underlying problems which demand different solutions from those at larger scales (38). If physicists and chemists already believe that to fully know the characteristics of a complex system—at the atomic or molecular level, may prove to be a rather difficult task, we must wonder what might be needed to deal with information spanning several more orders of magnitude in complexity, such as the historical, social, political, psychological, emotional and economic information that play such a significant role in biomedical informatics.
Researchers begin to face unique challenges that nano brings to informatics as referred to above—such as the polymorphic and polydisperse nature of nanomaterials. These characteristics imply a substantial added requirement for expert annotation and curation of data and analyses to inform scientists about the quality and reliability of the data, test methods, analyses, and models used in nanomedicine. Such challenges need to be addressed before semantic or ontological analyses, and may well influence the new area of translational nanoinformatics. That is, it will be use-driven in defining the information needed to advance the science and the translation to the clinic.
Education in the new area of nanoinformatics exacerbates the problems of range and depth of content and practice already faced by BMI. Nanoinformatics adds areas such as advanced quantum physics and chemistry, including nanotoxicity, with new models of imaging, and pharmacodynamics, which go beyond traditional research and curricula in BMI. Future professionals with expertise in nanoinformatics may have an important role to play as information brokers, connecting people with diverse backgrounds and expertise who would, otherwise, have a difficult time understanding each other. In such futuristic academic programs for nanoinformatics/nanomedicine, informatics methods and tools could play a central role for students and professionals by helping understand and manage the new concepts needed, without having to become quantum physicists or chemists.
In the above, we have focused on concrete applications of nanoparticles in nanomedicine. But, at the scientific level, there is a significant challenge related to the concept of “information.” The pioneering book by Blois (39) thoroughly explored the concept of information and its relevance to medicine, illustrating clearly the difficulties of defining information and giving a unified view of the field. A unified theory of biomedical information will require considerable advances, due to differences between bits and quantum bits, a measure of information at the quantum level; their appearance in nature and the diversity of meanings of the term “information” when used in different applications such as medical records, signals, images, scientific references, or news. Implications for medicine of novel scientific information theory may come from better understanding of information transmission in the brain, the mechanisms of consciousness or how information can be linked to 3-D molecular structures. These fundamental challenges in science are usually quite beyond mainstream research in BMI but will be essential to define the new informatics at the nano level.
Future Directions: DNA and RNA Computing
Recent advances in biotechnology and genetic engineering are allowing physicists, engineers, and computational scientists to explore the computational promise of biological mechanisms as a technology. Synthetic biology (40) and the engineering of biological systems (41) are new fields that consider a cell as a machine that can be programmed, controlled, regulated, and modulated. Specific genetic engineering tools will allow building biological hardware and software (biomolecular automata and genetic circuits, for example) that operate within a cell and help diagnose diseases and deliver treatments in vitro. In the near future, it is expected that these biomolecular devices will operate in vivo within living organisms.
Biomolecular computing (42) is the term used for information processing coded in biological macromolecules. A biomolecular computer is a device made with these biomolecules that processes biological information, and use DNA, RNA, proteins, or their combination. In this section, we will describe only a few of the most relevant DNA and RNA-based computers developed so far and applied to in vivo diagnostic and drug delivery.
Since 1994, when Adleman (43) solved a computational problem using biomolecules (DNA strands and enzymes), scientists have learned that biological computers will not compete with electronic computers in speed and accuracy for numerical solutions of conventional problems (popularly, number crunching). Their real potential lies in their natural role for operating in biochemical environments, sensing and analyzing biomolecular signals, and emitting biomolecular outputs. These biological and “wet-lab” environments are not hospitable for electronic devices but are perfect for biomolecular devices processing biological signals.
DNA-Based Automata
The engineering of programmable biomolecular automata applied to the diagnosis/treatment in vitro of a disease is a promising application in the area of intelligent in situ drug delivery. This field started in 2001 with the first design of a DNA-based automaton operating in vitro (44) applied to biomedical diagnosis in 2004 (45). An automaton is a device that can operate in an autonomous way, sensing inputs, processing those inputs and emitting an output without external human interaction.
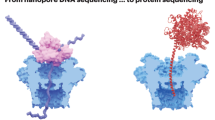
In the biomolecular automaton developed by Benenson and coworkers (45,46), the inputs of the device are molecular diagnostic biomarkers like messenger RNAs (mRNA). The computational unit (a double DNA strand called a diagnostic molecule in Fig. 3) processes the different inputs and emits a positive diagnosis output only if all the disease indicators are present. This computational unit has medical-diagnostic knowledge encoded in a chain of logical IF/THEN rules of the type: “IF mRNA_1 level is high THEN Diagnosis Positive.” The output in the case of a positive diagnosis will be the release of a single strand drug DNA molecule. This kind of automaton has two states: “Positive diagnosis” and “Negative diagnosis.” Only in the case that the automaton ends its transitions in the “Positive diagnosis” state will it deliver the drug DNA molecule. The transitions of the automata are executed with a restriction enzyme called Fok I that cuts the diagnostic molecule when processing each mRNA input. The cuts made by the Fok I enzyme in the two DNA strands are represented with arrows in Fig. 3.
Biomolecular automaton made with DNA strands and a Fok I restriction enzyme. The automaton will deliver a single stranded DNA drug molecule only if the input, an mRNA strand, is detected. Only in this situation, in the state of “Positive Diagnosis,” the enzyme Fok I will cut the diagnostic molecule to deliver the DNA drug. Adapted from Benenson Y et al., 2004; Nature 429:423–429; Copyright © 2004 Macmillan Publishers Ltd, with permission.
The Ehud Shapiro's group at the Weizmann Institute and other groups are working to solve the grand challenge of putting a “Doctor in a Cell”: a genetically modified cell that can operate in a human body, with a biological computer inside it that can process and analyze external biological signals, emit a diagnosis and deliver the desired molecular therapeutic signal (46).
DNA-Based Sensors and Circuits
Another important and widely used nucleic acid sensing technique is the so-called “competitive hybridization” or “strand displacement” (47). This technique is used in the design of DNA logic circuits (47,48) for the intelligent sensing and processing of DNA and RNA molecules.
The basic mechanism of a DNA or RNA sensor is based on competitive hybridization as shown in Fig. 4. The device detects the presence or absence of an input: the DNA (or RNA) strand called A. The sensor consist of two DNA strands B and A' partially hybridized. B and A' are not fully complementary. A' is the DNA strand complement of A. If the input A is present, it will displace B from the BA' complex due to it's stronger attraction for A' and the greater stability of the duplex AA'. The presence of A can be detected by attaching a fluorescent molecule at one end of B and a quencher molecule at the end of A'. The quencher molecule will avoid the fluorescence when A is not present. If A is present, B will emit fluorescence (Fig. 4). By using this simple technique sophisticated logic circuits detecting and analyzing complex patterns of DNA or RNA signals have been engineered (46,47).
(a) DNA sensor. A complex formed by two DNA strands B and A' can detect the presence of a DNA strand A by competitive hybridization between A and B. Only when A is present B strand will be displaced from the quencher in A' and will emit fluorescence. (b) RNA sensor. An RNA strand with a hairpin loop will change its conformational structure when it detects and binds to its ligand (chemical input). Only when the chemical input is present, the conformational change separates the fluorescent molecule from the quencher allowing the emission of fluorescence. Adapted from Davidson EA et al., 2007; Nat Chem Biol 3:23–28; Copyright © 2007 Macmillan Publishers Ltd., with permission.
RNA-Based Computers
New kinds of endogenous RNA molecules like antisense RNA, microRNA, small interference RNA (siRNA), riboregulators, ribozymes, and riboswitches with new genetic expression regulatory properties (basically, the ability to silence any precise and selected gene) have been discovered in the past 10 years. This process, called RNA interference (RNAi), was first explained in 1998 by the 2006 Nobel Laureates Fire and Mello (49). In 2002, RNAi was called by Science Journal as the “Breakthrough of the Year” for its potential to become a powerful therapeutic mechanism or drug. There are currently dozens of RNAi-based drug compounds in clinical trials. Excellent reviews covering RNA regulation activity and RNA synthetic biology can be found in Refs. 50,51.
The discovery of RNA interference has led to great interest in the development of new synthetic DNA and RNA-based computers that can sense, process and deliver these new RNA molecules. For example:
-
Win and Smolke (52) have developed an in vivo RNA system based on riboswitches and ribozymes. Riboswitches are RNA molecules that bind small ligands to regulate gene expression through conformational changes that control the expression activity (see the action of a simple riboswitch in Fig. 4b) and ribozymes are RNA enzymes that catalyze RNA cutting.
-
Benenson and coworkers (53) have developed a DNA logic circuit that can process and analyze up to five siRNA inputs. These siRNA are synthetic 20 nucleotide doubled-stranded RNAs that repress translation.
-
Winfree and coworkers (47) have developed logic circuits that can receive as inputs microRNAs (its function is similar to siRNA inhibiting protein translation through selective pairing with a target mRNA).
-
Bueno and Patón (54) have developed a synthetic device that generates oscillatory biomolecular signals using two interacting genes and one ribozyme.
Other relevant work includes Stojanovic's molecular automaton made with deoxyribozymes (55) and the pioneer works of Weiss and Knight (56) at MIT designing genetic circuits. In Ref. 57 Simmel describes biomedical applications of DNA devices and (58) contains a complete review of biocomputers. But, the grand challenge that faces the biomolecular computer engineering field is to design and follow a successful path from devices working in vitro to designs that can operate in vivo.
CONCLUSIONS
Various types of nanoparticles can be used in nanomedicine for different purposes. Dendrimers and nanospheres can be used as drug carriers or quantum dots can be used as markers for diagnosis and monitoring tasks. Research on quantum dots and the development of new nanoparticles for drug delivery benefit from physical characteristics that were not available previously, so nanoinformaticians deals with new challenges and will need different foundations and rationales than those that have been shown effective in the past at more macro levels.
From a software engineering perspective, there are many methods and tools that have been developed within BMI that could benefit nanomedicine. Informatics tools can be invaluable in accelerating the design and implementation of nanoparticles and devices and the evaluation of in vitro and in vivo applications, which will help advance nanomedicine. However, it is likely that the cost of research in nanomedicine will significantly increase the cost of research in biomedicine as the simulation and experimentation with particles, environments, functions, toxicities and other aspects of nanomedicine encourages more researchers to work on nanomedical applications. By creating new modeling and simulation methods and tools, building databases and ontologies and the interoperable infrastructures required for research; informatics researchers will have a decisive impact on nanomedicine, collaborating with advanced laboratories in academic settings, hospitals, and industry. Such informatics' research will require as much or more financial support than research is currently providing to BMI and systems biology, but is a natural extension of it, which increases the opportunities for advances in healthcare by many orders of magnitude.
We can anticipate that nanoparticles will have a primary role in medical research within the next decades and that nanomedicine will become part of routine medical care. From the perspective of national health systems, the increasing costs of new preventive approaches (biomarkers, routine genome sequencing, etc) and their application to entire populations may prove to be such a financial burden that support for personalized medicine will require new economic models for healthcare. Further costs arising from nanomedical technologies might deepen the existing divide between the rich and the poor both within and between nations, leading to even greater social and political problems. Yet, from a positive perspective, informatics may be able to support the investigation of planning and allocation strategies that could identify precisely those patients where the benefits of nanomedicine might prove most critical, cost effective, and of greatest human value.
Abbreviations
- BMI:
-
biomedical informatics
- NPO:
-
nanoparticle ontology
- NCI:
-
national cancer institute
References
Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM 2005 Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol 23: 1294–1301
Papagiannaros A, Levchenko T, Hartner W, Mongayt D, Torchilin V 2009 Quantum dots encapsulated in phospholipid micelles for imaging and quantification of tumors in the near-infrared region. Nanomedicine 5: 216–224
Leary SP, Liu CY, Apuzzo ML 2006 Toward the emergence of nanoneurosurgery: part II—nanomedicine: diagnostics and imaging at the nanoscale level. Neurosurgery 58: 805–823
Jain K 2008 The Handbook of Nanomedicine. Humana Press, New Jersey, pp 161–192
Freitas RA 2005 What is nanomedicine?. Nanomedicine 1: 2–9
Iyer AK, Khaled G, Fang J, Maeda H 2006 Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today 11: 812–818
De Jong WH, Borm PJ 2008 Drug delivery and nanoparticles: applications and hazards. Int J Nanomedicine 3: 133–149
Malam Y, Loizidou M, Seifalian AM 2009 Liposomes and nanoparticles: nanosized vehicles for drug delivery in cancer. Trends Pharmacol Sci 30: 592–599
Storhoff JJ, Marla SS, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, Cork W, Müller UR 2004 Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron 19: 875–883
Dufès C, Uchegbu IF, Schätzlein AG 2005 Dendrimers in gene delivery. Adv Drug Deliv Rev 57: 2177–2202
Ozdemir V, Williams-Jones B, Glatt SJ, Tsuang MT, Lohr JB, Reist C 2006 Shifting emphasis from pharmacogenomics to theragnostics. Nat Biotechnol 24: 942–946
Harisinghani MG, Barentsz J, Hahn PF, Deserno WM, Tabatabaei S, van de Kaa CH, de la Rosette J, Weissleder R 2003 Noninvasive detection of clinically occult lymph-node metastases in prostate cancer. N Engl J Med 348: 2491–2499
Gradishar WJ 2006 Albumin-bound paclitaxel: a next-generation taxane. Expert Opin Pharmacother 7: 1041–1053
Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA 2008 Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359: 366–377
Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M 2007 Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450: 1235–1239
Frey LJ, Maojo V, Mitchell JA 2007 Bioinformatics linkage of heterogeneous clinical and genomic information in support of personalized medicine. Yearb Med Inform: 98–105. Erratum in: Yearb Med Inform 2008: 19, 2007
McCabe ER 2010 nanopediatrics: enabling personalized medicine for children. Pediatr Res 67: 453–457
Marin S, Merkoci 2009 A direct electrochemical stripping detection of cystic-fibrosis-related DNA linked through cadmium sulfide quantum dots. Nanotechnology 20: 55101
Mortensen NP, Fowlkes JD, Sullivan CJ, Allison DP, Larsen NB, Molin S, Doktycz MJ 2009 Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25: 3728–3733
Dames P, Gleich B, Flemmer A, Hajek K, Seidl N, Wiekhorst F, Eberbeck D, Bittmann I, Bergemann C, Weyh T, Trahms L, Rosenecker J, Rudolph C 2007 Targeted delivery of magnetic aerosol droplets to the lung. Nat Nanotechnol 2: 495–499
Stone V, Johnston H, Clift MJ 2007 Air pollution, ultrafine and nanoparticle toxicology: cellular and molecular interactions. IEEE Trans Nanobioscience 6: 331–340
Dobson J 2007 Toxicological aspects and applications of nanoparticles in paediatric respiratory disease. Paediatr Respir Rev 8: 62–66
Service RF 2004 Nanotoxicology. Nanotechnology grows up. Science 304: 1732–1734
Anguita A, Martín L, Crespo J, Tsiknakis M 2008 An ontology-based method to solve query identifier heterogeneity in post-genomic clinical trials. Stud Health Technol Inform 136: 3–8
McCusker JP, Phillips JA, Beltrán AG, Finkelstein A, Krauthammer M 2009 Semantic web data warehousing for caGrid. BMC Bioinformatics 10: S2
Alonso-Calvo R, Maojo V, Billhardt H, Martin-Sanchez F, García-Remesal M, Perez-Rey D 2007 An agent- and ontology-based system for integrating public gene, protein, and disease databases. J Biomed Inform 40: 17–29
Perez-Rey D, Maojo V, García-Remesal M, Alonso-Calvo R, Billhardt H, Martin-Sanchez F, Sousa A 2006 ONTOFUSION: ontology-based integration of genomic and clinical databases. Comput Biol Med 36: 712–730
Tomalia DA 2009 In quest of a systematic framework for unifying and defining nanoscience. J Nanopart Res 11: 1251–1310
Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey MA, Chute CG, Musen MA 2009 BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res 37 (Web Server issue): W170–W173
Brazas MD, Fox JA, Brown T, McMillan S, Ouellette BF 2008 Keeping pace with the data: 2008 update on the bioinformatics links directory. Nucleic Acids Res 36 (Web Server issue): W2–W4
de la Calle G, García-Remesal M, Chiesa S, de la Iglesia D, Maojo V 2009 BIRI: a new approach for automatically discovering and indexing available public bioinformatics resources from the literature. BMC Bioinformatics 10: 320–328
Chiesa S, García-Remesal M, de la Calle G, de la Iglesia D, Bankauskaite V, Maojo V 2010 Building an index of nanomedical resources: an automatic approach based on text mining. In: Lovrek I, Howlett RJ, Jain LC (eds) Knowledge-Based Intelligent Information and Engineering Systems. Springer-Verlag, Berlin, pp 50–57
Chiesa S, De La Iglesia D, Crespo J, Martin-Sanchez F, Kern J, Potamias G, Maojo V 2009 European efforts in nanoinformatics research applied to nanomedicine. Stud Health Technol Inform 150: 757–761
Brazma A 2009 Minimum Information About a Microarray Experiment (MIAME)–successes, failures, challenges. Scientific WorldJournal 9: 420–423
Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Consortium OB, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S 2007 The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25: 1251–1255
Thomas DG, Pappu RV, Baker NA 2009 Ontologies for cancer nanotechnology research. Conf Proc IEEE Eng Med Biol Soc 1: 4158–4161
Gowers T, Nielsen M 2009 Massively collaborative mathematics. Nature 461: 879–881
Roukes M 2001 Plenty of room, indeed. Sci Am 285: 48–51: 54-57
Blois MS 1994 Information and Medicine: The Nature of Medical Descriptions. University of California Press, Berkeley, CA, pp 1–37
Purnick PE, Weiss R 2009 The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol 10: 410–422
Endy D 2005 Foundations for engineering biology. Nature 438: 449–453
Amos M 2005 Models of molecular computation. Theoretical and Experimental DNA Computation. Springer-Verlag, Berlin, pp 45–70
Adleman LM 1994 Molecular computation of solutions to combinatorial problems. Science 266: 1021–1024
Benenson Y, Paz-Elitzur T, Adar R, Keinan E, Livneh Z, Shapiro E 2001 Programmable and autonomous computing machine made of biomolecules. Nature 414: 430–434
Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E 2004 An autonomous molecular computer for logical control of gene expression. Nature 429: 423–429
Shapiro E, Benenson Y 2006 Tapping the computing power of biological molecules gives rise to tiny machines that can speak directly to living cells. Sci Am 295: 44–51
Seelig G, Soloveichik D, Zhang DY, Winfree E 2006 Enzyme-free nucleic acid logic circuits. Science 314: 1585–1588
Zhang DY, Turberfield AJ, Yurke B, Winfree E 2007 Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318: 1121–1125
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998 Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811
Isaacs FJ, Dwyer DJ, Collins JJ 2006 RNA synthetic biology. Nat Biotechnol 24: 545–554
Davidson EA, Ellington AD 2007 Synthetic RNA circuits. Nat Chem Biol 3: 23–28
Win MN, Smolke CD 2008 Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460
Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y 2007 A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol 25: 795–801
Miró Bueno JM, Rodríguez-Patón A 2009 A new model of synthetic genetic oscillator based on trans-acting repressor ribozyme. In: Omatu S, Rocha MP, Bravo J, Fernández-Riverola F, Corchado E, Bustillo A, Corchado JM (eds) Distributed Computing, Artificial Intelligence, Bioinformatics, Soft Computing, and Ambient Assisted Living. Springer-Verlag, Berlin, pp 1170–1177
Stojanovic MN, Stefanovic D 2003 A deoxyribozyme-based molecular automaton. Nat Biotechnol 21: 1069–1074
Weiss R, Homsy GE, Knight TF 2002 Toward in vivo digital circuits. In: Landweber LF, Winfree E (eds) Evolution as Computation: DIMACS Workshop. Springer-Verlag, Berlin, pp 275–295
Simmel FC 2007 Towards biomedical applications for nucleic acid nanodevices. Nanomedicine 2: 817–830
Benenson Y 2009 Biocomputers: from test tubes to live cells. Mol Biosyst 5: 675–685
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported, in part, by the European Commission (Action-Grid project), the Ministry of Science and Innovation of Spain (COMBIOMED RETICS, FIS and the ONTOMINEBASE project), and the Comunidad de Madrid.
Rights and permissions
About this article
Cite this article
Maojo, V., Martin-Sanchez, F., Kulikowski, C. et al. Nanoinformatics and DNA-Based Computing: Catalyzing Nanomedicine. Pediatr Res 67, 481–489 (2010). https://doi.org/10.1203/PDR.0b013e3181d6245e
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e3181d6245e
This article is cited by
-
A Minor Groove Binder with Significant Cytotoxicity on Human Lung Cancer Cells: The Potential of Hesperetin Functionalised Silver Nanoparticles
Journal of Fluorescence (2023)
-
Nanoinformatics and biomolecular nanomodeling: a novel move en route for effective cancer treatment
Environmental Science and Pollution Research (2020)
-
High-performance biosensing based on autonomous enzyme-free DNA circuits
Topics in Current Chemistry (2020)