Abstract
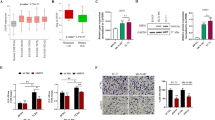
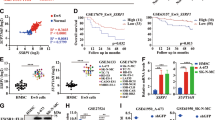
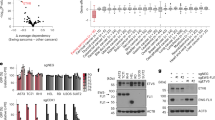
Ewing Sarcoma is a biologically aggressive bone and soft tissue malignancy affecting children and young adults. Ewing Sarcoma pathogenesis is driven by EWS/Ets fusion oncoproteins, of which EWS/Fli1 is the most common. We have previously shown that microRNAs (miRs) regulated by EWS/Fli1 contribute to the pro-oncogenic program in Ewing Sarcoma. Here we show that miR-22, an EWS/Fli1-repressed miR, is inhibitory to Ewing Sarcoma clonogenic and anchorage-independent cell growth, even at modest overexpression levels. Our studies further identify the H3K9me1/2 histone demethylase KDM3A (JMJD1A/JHDM2A) as a new miR-22-regulated gene. We show that KDM3A is overexpressed in Ewing Sarcoma, and that its depletion inhibits clonogenic and anchorage-independent growth in multiple patient-derived cell lines, and tumorigenesis in a xenograft model. KDM3A depletion further results in augmentation of the levels of the repressive H3K9me2 histone mark, and downregulation of pro-oncogenic factors in Ewing Sarcoma. Together, our studies identify the histone demethylase KDM3A as a new, miR-regulated, tumor promoter in Ewing Sarcoma.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ludwig JA . Ewing sarcoma: historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr Opin Oncol 2008; 20: 412–418.
Toomey EC, Schiffman JD, Lessnick SL . Recent advances in the molecular pathogenesis of Ewing's sarcoma. Oncogene 2010; 29: 4504–4516.
Ghildiyal M, Zamore PD . Small silencing RNAs: an expanding universe. Nat Rev Genet 2009; 10: 94–108.
Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z . Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 2009; 15: 1443–1461.
Kasinski AL, Slack FJ . Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011; 11: 849–864.
McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK et al. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene 2011; 30: 4910–4920.
Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN et al. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing's sarcoma. Oncogene 2011; 30: 2173–2180.
De Vito C, Riggi N, Suva ML, Janiszewska M, Horlbeck J, Baumer K et al. Let-7a is a direct EWS-FLI-1 target implicated in Ewing's sarcoma development. PLoS One 2011; 6: e23592.
Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O . MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene 2012; 32: 3915–3921.
Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S et al. miR-34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol 2012; 226: 796–805.
Baylin SB, Jones PA . A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer 2011; 11: 726–734.
Ryan RJ, Bernstein BE . Molecular biology. Genetic events that shape the cancer epigenome. Science 2012; 336: 1513–1514.
Lawlor ER, Thiele CJ . Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res 2012; 18: 2768–2779.
Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 2012; 481: 329–334.
Li J, Liang S, Yu H, Zhang J, Ma D, Lu X . An inhibitory effect of miR-22 on cell migration and invasion in ovarian cancer. Gynecol Oncol 2010; 119: 543–548.
Li J, Zhang Y, Zhao J, Kong F, Chen Y . Overexpression of miR-22 reverses paclitaxel-induced chemoresistance through activation of PTEN signaling in p53-mutated colon cancer cells. Mol Cell Biochem 2011; 357: 31–38.
Nagaraja AK, Creighton CJ, Yu Z, Zhu H, Gunaratne PH, Reid JG et al. A link between mir-100 and FRAP1/mTOR in clear cell ovarian cancer. Mol Endocrinol 2010; 24: 447–463.
Patel JB, Appaiah HN, Burnett RM, Bhat-Nakshatri P, Wang G, Mehta R et al. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene 2011; 30: 1290–1301.
Ting Y, Medina DJ, Strair RK, Schaar DG . Differentiation-associated miR-22 represses Max expression and inhibits cell cycle progression. Biochem Biophys Res Commun 2010; 394: 606–611.
Xiong J, Du Q, Liang Z . Tumor-suppressive microRNA-22 inhibits the transcription of E-box-containing c-Myc target genes by silencing c-Myc binding protein. Oncogene 2010; 29: 4980–4988.
Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J 2010; 277: 1684–1694.
Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S et al. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer 2010; 103: 1215–1220.
Gurtan AM, Sharp PA . The role of miRNAs in regulating gene expression networks. J Mol Biol 2013; 425: 3582–3600.
Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C et al. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 2005; 65: 11459–11468.
Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res 2008; 68: 2176–2185.
Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O . Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007; 11: 421–429.
Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006; 125: 483–495.
Cloos PA, Christensen J, Agger K, Helin K . Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 2008; 22: 1115–1140.
Li B, Carey M, Workman JL . The role of chromatin during transcription. Cell 2007; 128: 707–719.
Wai DH, Schaefer KL, Schramm A, Korsching E, Van Valen F, Ozaki T et al. Expression analysis of pediatric solid tumor cell lines using oligonucleotide microarrays. Int J Oncol 2002; 20: 441–451.
Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer 2012; 131: E179–E189.
Chen H, Kluz T, Zhang R, Costa M . Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis 2010; 31: 2136–2144.
Mimura I, Nangaku M, Kanki Y, Tsutsumi S, Inoue T, Kohro T et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol 2012; 32: 3018–3032.
Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci USA 2009; 106: 5324–5329.
Douglas D, Hsu JH, Hung L, Cooper A, Abdueva D, van Doorninck J et al. BMI-1 promotes ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res 2008; 68: 6507–6515.
Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res 2012; 22: 259–270.
Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011; 331: 435–439.
Geutjes EJ, Bajpe PK, Bernards R . Targeting the epigenome for treatment of cancer. Oncogene 2012; 31: 3827–3844.
Erkizan HV, Kong Y, Merchant M, Schlottmann S, Barber-Rotenberg JS, Yuan L et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma. Nat Med 2009; 15: 750–756.
Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst 2011; 103: 962–978.
Greer EL, Shi Y . Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–357.
Fukuma M, Okita H, Hata J, Umezawa A . Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene 2003; 22: 1–9.
Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M et al. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem 2009; 284: 733–739.
Cittelly DM, Dimitrova I, Howe EN, Cochrane DR, Jean A, Spoelstra NS et al. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Mol Cancer Ther 2012; 11: 2556–2565.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307: 1098–1101.
Acknowledgements
We thank Kathrin Bernt and Tobias Neff for advice on histone mark analysis, Steve Lessnick for retroviral packaging constructs, Heide Ford for critical reading of the manuscript, and the University of Colorado Cancer Center DNA Sequencing, Flow Cytometry, and Functional Genomics core facilities. This work was supported by the Boettcher Foundation’s Webb-Waring Biomedical Research Program, Department of Defense Discovery Award (W81XWH-12-1-0296), Alex’s Lemonade Stand Foundation for Childhood Cancer, and funds from the University of Colorado School of Medicine and Cancer Center (to PJ); and Merit Award from the US Department of Veterans’ Affairs and R01CA138528-2522717 (to RAW).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Parrish, J., Sechler, M., Winn, R. et al. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene 34, 257–262 (2015). https://doi.org/10.1038/onc.2013.541
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.541
Keywords
This article is cited by
-
KDM3A-mediated SP1 activates PFKFB4 transcription to promote aerobic glycolysis in osteosarcoma and augment tumor development
BMC Cancer (2022)
-
Histone demethylase JMJD1A promotes expression of DNA repair factors and radio-resistance of prostate cancer cells
Cell Death & Disease (2020)
-
miRNA signatures in childhood sarcomas and their clinical implications
Clinical and Translational Oncology (2019)
-
The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis
Oncogene (2017)
-
Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival
Oncogene (2016)