Abstract
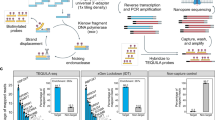
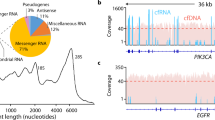
Most human cancers are characterized by genetic aberrations accompanied by altered expression and function of numerous genes. Applying genome-wide, microarray gene expression analysis to identify deregulated genes in different tumour types can provide potential gene candidates as diagnostic and prognostic tools and promising targets for drug development. However, the detection of deregulated genes with low levels of expression remains a major challenge. In this study, we have designed a strategy, termed modified suppression subtractive hybridization (mSSH), to identify genes encoding rare transcripts. The strategy entails incorporating the T7-promoter sequence at the 5′ end of the noncoding cDNA strand during first strand cDNA synthesis to generate unidirectional antisense RNA from the resultant cDNA following conventional SSH. These transcripts are subsequently analysed by Affymetrix oligonucleotide gene arrays. Here, we have used five hepatocellular carcinoma (HCC), five breast carcinoma and four nasopharyngeal carcinoma (NPC) biopsies separately as testers and their corresponding normal biopsies as drivers to enrich for low abundance tumour type-specific transcripts. The total detectable number of probe sets following mSSH was reduced almost 10-fold in comparison to those detected for the same resected tumour tissues without undergoing subtraction, thus yielding a subtraction efficacy of over 90%. Using this experimental approach, we have identified 48 HCC-specific, 45 breast carcinoma-specific, and 83 NPC-specific genes. In addition, 115 genes were upregulated in all the three cancer types. When compared to gene-profiling data obtained without mSSH, the majority of these identified transcripts were of low abundance in the original cancer tissues. mSSH can therefore serve as a comprehensive molecular strategy for pursuing functional genomic studies of human cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. (2000). Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511.
Amatscheck S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A et al. (2004). Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res 64: 844–856.
Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE et al. (2002). Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8: 816–824.
Bonavida B . (2007). Rituximab-induced inhibition of antiapoptotic cell survival pathways: impolications in chemo/immunoresistance, rituximab unresponsiveness, prognostic and novel therapeutic interventions. Oncogene 26: 3629–3636.
Brentani H, Caballero OL, Camargo AA, Silva AM, Silva Jr WA, Neto ED et al. (2003). The generation and utilization of a cancer-oriented representation of the human transcriptome by using expressed sequence tags. Proc Natl Acad Sci USA 100: 13418–13423.
Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A et al. (2007). A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 356: 11–20.
Comijn J, Berx G, Vermassen P, Verschueren K, Grunsven L, Bruyneel E et al. (2001). The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasions. Mol Cell 7: 1267–1278.
Del Rio M, Molina F, Bascoul-Mollevi C, Copois V, Bibeau F, Chalbos P et al. (2007). Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol 25: 773–780.
Diatchenko L, Lau YC, Campbell AP, Chenchik A, Moqadam F, Huang B et al. (1996). Suppresion subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libralies. Proc Natl Acad Sci USA 93: 6025–6030.
Draghici S, Khatri P, Eklund AC, Szallasi Z . (2006). Reliability and reproducibility issues in DNA microarray measurements. Trends Genet 22: 101–109.
Kirk GD, Lesi OA, Mendy M, Szymanska K, Whittle H, Goedert JJ et al. (2005). 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene 24: 5858–5867.
Lah TT, Cercek M, Blejee A, Kos J, Gorodetsky E, Somers R et al. (2000). Cathepsin B, a prognostic indicator in lymp node-negative breast carcinoma patients: comparison with Cathepsin D, Cathepsin L, and other clinical indicators. Clin Cancer Res 6: 578–584.
Lau WM, Ho TH, Hui KM . (2007). p16(INK4A)-silencing augments DNA damage-induced apoptosis in cervical cancer cells. Oncogene 26: 6050–6060.
Lee SW, Tomasetto C, Sager R . (1991). Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc Natl Acad Sci USA 88: 2825–2829.
Lipshutz RJ, Fodor SPA, Gingeras TR, Lockhart DJ . (1999). High density synthetic oligonucleotide arrays. Nat Genet 21: 20–24.
Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL et al. (2004). Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378–1381.
Qin LX, Beyer RP, Hudson FN, Linford NJ, Morris DE, Kerr KF . (2006). Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics 7: 23.
Russo G, Zegar C, Goirdano A . (2003). Advantages and limitations of micoarray technology in human cancer. Oncogene 22: 6497–6507.
Sander C . (2000). Genomic medicine and the future of health care. Science 287: 1977–1978.
Shiota G, Kishimoto Y, Suyama A, Okubo M, Katayama S, Harada K et al. (1997). Prognostic significance of serum anti-p53 antibody in patients with hepatocellular carcinoma. J Hepatol 27: 661–668.
Thornburg NJ, Kulwichit W, Edwards RH, Shair KHY, Bendt KM, Raab-Traub N . (2006). LMP1 signalling and activation of NF-kB in LMP1 transgenic mice. Oncogene 25: 288–297.
Thuerigen O, Schneeweiss A, Toedt G, Warnat P, Hahn M, Kramer H et al. (2006). Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J Clin Oncol 20: 1839–1845.
Tsopanomichalou M, Kouroumalis E, Ergazaki M, Spandidos DA . (1999). Loss of heterozygosity and microsatellite instability in human non-neoplastic hepatic lesions. Liver 19: 305–311.
Wang S, Ooi L, Hui KM . (2007). Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res 13: 6275–6283.
Young LS, Murray PG . (2003). Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22: 5108–5121.
Young LS, Rickinson AB . (2004). Epstein-Barr virus: 40 years on. Nat Rev Cancer 4: 757–768.
Yu K, Kumaresan G, Miller LD, Tan P . (2006). A modular analysis of breast cancer reveals a novel low-grade molecular signature in estrogen receptor-positive tumors. Clin Cancer Res 12: 3288–3296.
Acknowledgements
We greatly appreciate the critical reading of this manuscript by our colleague Professor Malcolm Paterson and Mr Yu Kun for his assistance the statistical analyses. This work was supported by grants from the National Medical Research Council of Singapore.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary information
Rights and permissions
About this article
Cite this article
Liu, B., Goh, C., Ooi, L. et al. Identification of unique and common low abundance tumour-specific transcripts by suppression subtractive hybridization and oligonucleotide probe array analysis. Oncogene 27, 4128–4136 (2008). https://doi.org/10.1038/onc.2008.50
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2008.50
Keywords
This article is cited by
-
The molecular underpinning of geminin-overexpressing triple-negative breast cancer cells homing specifically to lungs
Cancer Gene Therapy (2022)
-
IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis
Molecular Cancer (2017)
-
Differentially expressed genes between female and male adult Anopheles anthropophagus
Parasitology Research (2009)