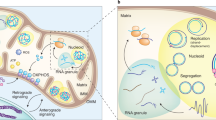
Key Points
-
Mitochondrial DNA is essential, but for many years mammalian mitochondria were thought to lack repair systems for their DNA. Now it is well established that base excision repair serves a key role in maintaining the integrity of the mitochondrial genome, and factors involved in all of the other major repair pathways have been assigned to mitochondria, albeit with differing degrees of confidence.
-
Much of the past, and indeed present, confusion arose from the fact that many mitochondrial DNA repair proteins are shared with the nucleus, and so the initial goal in assessing a protein's contribution to mitochondrial DNA repair is to establish it as a bona fide mitochondrial resident, and not merely a contaminant. Mitochondrial and nuclear DNA repair proteins that are encoded by the same gene can often be distinguished owing to the presence or absence of a mitochondrial targeting signal. Signal sequences may be generated through alternative splicing, alternative transcription initiation or alternative translation initiation.
-
The machinery of mitochondrial base excision repair overlaps considerably with that of the nucleus. By contrast, mismatch repair, a more recently accepted part of the DNA repair armoury, is less reliant on dually localized proteins.
-
The procedure for dealing with single-strand breaks in mitochondrial DNA is becoming clear, but there are still many gaps in our knowledge regarding mitochondrial double-strand break repair.
-
Human diseases and genetic manipulations are providing a wealth of new data on the factors contributing to mitochondrial DNA integrity, aiding the building of a comprehensive inventory of mitochondrial DNA repair factors.
-
As in other systems, mitochondrial DNA repair is likely to be integrated with the sister processes of replication and recombination. Aberrations in any or all these processes can give rise to pathological forms of mitochondrial DNA.
Abstract
Mitochondrial DNA (mtDNA) faces the universal challenges of genome maintenance: the accurate replication, transmission and preservation of its integrity throughout the life of the organism. Although mtDNA was originally thought to lack DNA repair activity, four decades of research on mitochondria have revealed multiple mtDNA repair pathways, including base excision repair, single-strand break repair, mismatch repair and possibly homologous recombination. These mtDNA repair pathways are mediated by enzymes that are similar in activity to those operating in the nucleus, and in all cases identified so far in mammals, they are encoded by nuclear genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
03 October 2012
In the above article, the sentence “This study was the first to identify a protein (YB1) involved in mitochondrial MMR” incorrectly appeared below reference 107 instead of below reference 103 (de Souza-Pinto, N. C. et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair 8, 704–719 (2009)). This has been corrected online. The authors apologize for any confusion caused to readers.
References
Trifunovic, A. et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 (2004). This report shows that overwhelming the DNA repair capacity of mitochondria leads to multisystem disease and premature ageing.
Cerritelli, S. M. et al. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Biol. Rep. 11, 807–815 (2003).
Simsek, D. et al. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471, 245–248 (2011).
Li, Y. et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nature Genet. 11, 376–381 (1995).
Lan, L. et al. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA 101, 13738–13743 (2004).
Szczesny, B., Tann, A. W., Longley, M. J., Copeland, W. C. & Mitra, S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 283, 26349–26356 (2008). One of the first reports documenting mitochondrial LP-BER.
Pinz, K. G. & Bogenhagen, D. F. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol. Cell. Biol. 18, 1257–1265 (1998).
Stucki, M. et al. Mammalian base excision repair by DNA polymerases δ and ε. Oncogene 17, 835–843 (1998).
Hegde, M. L., Hazra, T. K. & Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 18, 27–47 (2008).
Campalans, A., Amouroux, R., Bravard, A., Epe, B. & Radicella, J. P. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J. Cell Sci. 120, 23–32 (2007).
Stuart, J. A., Mayard, S., Hashiguchi, K., Souza-Pinto, N. C. & Bohr, V. A. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 33, 3722–3732 (2005).
Boesch, P., Ibrahim, N., Dietrich, A. & Lightowlers, R. N. Membrane association of mitochondrial DNA facilitates base excision repair in mammalian mitochondria. Nucleic Acids Res. 38, 1478–1488 (2010).
He, J. et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 176, 141–146 (2007).
Chen, L., Haushalter, K. A., Lieber, C. M. & Verdine, G. L. Direct visualization of a DNA glycosylase searching for damage. Chem. Biol. 9, 345–350 (2002).
Slupphaug, G. et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92 (1996). This study demonstrated that mitochondrial and nuclear isoforms of a mammalian DNA repair enzyme derive from a single gene.
Demple, B. & Linn, S. 5,6-Saturated thymine lesions in DNA: production by ultraviolet light or hydrogen peroxide. Nucleic Acids Res. 10, 3781–3789 (1982).
Kao, J. Y., Goljer, I., Phan, T. A. & Bolton, P. H. Characterization of the effects of a thymine glycol residue on the structure, dynamics, and stability of duplex DNA by NMR. J. Biol. Chem. 268, 17787–17793 (1993).
Lipscomb, L. A. et al. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA 92, 719–723 (1995).
Dunn, A. R., Kad, N. M., Nelson, S. R., Warshaw, D. M. & Wallace, S. S. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 39, 7487–7498 (2011).
Slupphaug, G. et al. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 21, 2579–2584 (1993).
Nilsen, H. et al. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 25, 750–755 (1997).
Nakabeppu, Y. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative DNA damage. Prog. Nucleic Acid. Res. Mol. Biol. 68, 75–94 (2001).
Demple, B. & Sung, J.-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 4, 1442–1449 (2005).
Ikeda, S., Kohmoto, T., Tabata, R. & Seki, Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA Repair 1, 847–854 (2002).
Demple, B. & Harrison, L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63, 915–948 (1994).
Dou, H. et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J. Biol. Chem. 283, 3130–3140 (2008).
Waga, S. & Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 (1998).
Hazra, T. K. et al. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair 6, 470–480 (2007).
Hazra, T. K. et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA 99, 3523–3528 (2002).
Dou, H., Mitra, S. & Hazra, T. K. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 (2003).
Das, A. et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair 5, 1439–1448 (2006).
Wiederhold, L. et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220 (2004).
Mandal, S. M. et al. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3′-phosphatase in maintenance of mitochondrial genome. J. Biol. Chem. 287, 2819–2829 (2012).
Tahbaz, N., Subedi, S. & Weinfeld, M. Role of polynucleotide kinase/phosphatase in mitochondrial DNA repair. Nucleic Acids Res. 40, 3484–3495 (2011).
Pascucci, B., Russo, M. T., Crescenzi, M., Bignami, M. & Dogliotti, E. The accumulation of MMS-induced single strand breaks in G1 phase is recombinogenic in DNA polymerase β defective mammalian cells. Nucleic Acids Res. 33, 280–288 (2005).
Nielsen-Preiss, S. M. & Low, R. L. Identification of a β-like DNA polymerase activity in bovine heart mitochondria. Arch. Biochem. Biophys. 374, 229–240 (2000).
Longley, M. J., Prasad, R., Srivastava, D. K., Wilson, S. H. & Copeland, W. C. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase γ and its role in mitochondrial base excision repair in vitro. Proc. Natl Acad. Sci. USA 95, 12244–12248 (1998). This study identified 5′ dRP lyase activity of Pol γ.
Pinz, K. G. & Bogenhagen, D. F. The influence of the DNA polymerase γ accessory subunit on base excision repair by the catalytic subunit. DNA Repair 5, 121–128 (2006). One of the early reports documenting mitochondrial SP-BER.
Martin, I. V. & MacNeill, S. A. ATP-dependent DNA ligases. Genome Biol. 3, REVIEWS3005 (2002).
Liu, P. et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 28, 4975–4987 (2008).
Ischenko, A. A. & Saparbaev, M. K. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415, 183–187 (2002).
Pinz, K. G. & Bogenhagen, D. F. Characterization of a catalytically slow AP lyase activity in DNA polymerase γ and other family A DNA polymerases. J. Biol. Chem. 275, 12509–12514 (2000).
Garg, P., Stith, C. M., Sabouri, N., Johansson, E. & Burgers, P. M. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 18, 2764–2773 (2004).
Stewart, J. A., Campbell, J. L. & Bambara, R. A. Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 284, 8283–8291 (2009).
Bae, S. H., Bae, K. H., Kim, J. A. & Seo, Y. S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 (2001).
Duxin, J. P. et al. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 29, 4274–4282 (2009).
Zheng, L. et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32, 325–336 (2008).
Kang, H. Y. et al. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that Dna2 plays an essential role in Okazaki fragment metabolism. Genetics 155, 1055–1067 (2000).
Kalifa, L., Beutner, G., Phadnis, N., Sheu, S. & Sia, E. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair 8, 1242–1249 (2009).
Budd, M. E. & Campbell, J. L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17, 2136–2142 (1997).
Akbari, M., Visnes, T., Krokan, H. E. & Otterlei, M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair 7, 605–616 (2008).
Tann, A. W. et al. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 286, 31975–31983 (2011). Provocatively, this paper suggests that EXOG, rather than FEN1 and DNA2, mediates LP-BER in mitochondria.
Lakshmipathy, U. & Campbell, C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 28, 3880–3886 (2000).
Achanta, G. et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol γ. EMBO J. 24, 3482–3492 (2005).
Mummenbrauer, T. et al. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell 85, 1089–1099 (1996).
de Souza-Pinto, N. C., Harris, C. C. & Bohr, V. A. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene 23, 6559–6568 (2004).
Chen, D., Yu, Z., Zhu, Z. & Lopez, C. D. The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res. 66, 3485–3494 (2006).
De, S. et al. RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 125, 2509–2522 (2012).
Kamenisch, Y. et al. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J. Exp. Med. 207, 379–390 (2010).
Canugovi, C. et al. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair 9, 1080–1089 (2010).
Huang, J. C., Zamble, D. B., Reardon, J. T., Lippard, S. J. & Sancar, A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc. Natl Acad. Sci. USA 91, 10394–10398 (1994).
Reeves, R. & Adair, J. E. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair 4, 926–938 (2005).
Pohjoismaki, J. L. et al. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 34, 5815–5828 (2006).
Muller, T. A., Meek, K. & Hausinger, R. P. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair 9, 58–65 (2010).
Westbye, M. P. et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 283, 25046–25056 (2008).
Wollen Steen, K. et al. mtSSB may sequester UNG1 at mitochondrial ssDNA and delay uracil processing until the dsDNA conformation is restored. DNA Repair 11, 82–91 (2012).
Vartanian, V. et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl Acad. Sci. USA 103, 1864–1869 (2006).
Sengupta, S., Mantha, A. K., Mitra, S. & Bhakat, K. K. Human AP endonuclease (APE1/Ref-1) and its acetylation regulate YB-1-p300 recruitment and RNA polymerase II loading in the drug-induced activation of multidrug resistance gene MDR1. Oncogene 30, 482–493 (2011).
Holt, I. J. Mitochondrial DNA replication and repair: all a flap. Trends Biochem. Sci. 34, 358–365 (2009).
Caldecott, K. W. Single-strand break repair and genetic disease. Nature Rev. Genet. 9, 619–631 (2008).
Sykora, P., Wilson, D. M. & Bohr, V. A. Repair of persistent strand breaks in the mitochondrial genome. Mech. Ageing Dev. 133, 169–175 (2012).
Rossi, M. N. et al. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 284, 31616–31624 (2009).
de Murcia, J. M. et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA 94, 7303–7307 (1997).
Wang, Z. Q. et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11, 2347–2358 (1997).
Whatcott, C. J., Meyer-Ficca, M. L., Meyer, R. G. & Jacobson, M. K. A specific isoform of poly(ADP-ribose) glycohydrolase is targeted to the mitochondrial matrix by a N-terminal mitochondrial targeting sequence. Exp. Cell Res. 315, 3477–3485 (2009).
Niere, M. et al. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose). J. Biol. Chem. 287, 16088–16102 (2012).
Das, B. B., Dexheimer, T. S., Maddali, K. & Pommier, Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl Acad. Sci. USA 107, 19790–19795 (2010).
Das, B., Dexheimer, T., Maddali, K. & Pommier, Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl Acad. Sci. USA 107, 19790–19795 (2010).
Ahel, I. et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443, 713–716 (2006).
Sykora, P., Croteau, D. L., Bohr, V. A. & Wilson, D. M. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl Acad. Sci. USA 108, 7437–7442 (2011).
Stracker, T. H. & Petrini, J. H. J. The MRE11 complex: starting from the ends. Nature Rev. Mol. Cell Biol. 12, 90–103 (2011).
Bacman, S. R., Williams, S. L. & Moraes, C. T. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 37, 4218–4226 (2009). This study provides clear, albeit indirect, evidence of mitochondrial DSB repair in vivo.
Symington, L. S. & Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011).
Dmitrieva, N. I., Malide, D. & Burg, M. B. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R632–R640 (2011).
Kalifa, L. et al. Mitochondrial genome maintenance: roles for nuclear nonhomologous end-joining proteins in Saccharomyces cerevisiae. Genetics 190, 951–964 (2012).
Papeta, N. et al. Prkdc participates in mitochondrial genome maintenance and prevents Adriamycin-induced nephropathy in mice. J. Clin. Invest. 120, 4055–4064 (2010).
Sage, J. M., Gildemeister, O. S. & Knight, K. L. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 285, 18984–18990 (2010).
Kleff, S., Kemper, B. & Sternglanz, R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 11, 699–704 (1992).
Minczuk, M. et al. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 39, 4284–4299 (2011).
Thyagarajan, B., Padua, R. A. & Campbell, C. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 271, 27536–27543 (1996).
Holt, I. J., Dunbar, D. R. & Jacobs, H. T. Behaviour of a population of partially duplicated mitochondrial DNA molecules in cell culture: segregation, maintenance and recombination dependent upon nuclear background. Hum. Mol. Genet. 6, 1251–1260 (1997).
D'Aurelio, M. et al. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13, 3171–3179 (2004).
Gilkerson, R. W., Schon, E. A., Hernandez, E. & Davidson, M. M. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell Biol. 181, 1117–1128 (2008).
Grainge, I., Lesterlin, C. & Sherratt, D. J. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 39, 5140–5148 (2011).
Hyvarinen, A. K. et al. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 35, 6458–6474 (2007).
Hyvarinen, A. K., Pohjoismaki, J. L., Holt, I. J. & Jacobs, H. T. Overexpression of MTERFD1 or MTERFD3 impairs the completion of mitochondrial DNA replication. Mol. Biol. Rep. 38, 1321–1328 (2011).
Bailey, L. J. et al. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 37, 2327–2335 (2009).
Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981).
Mita, S. et al. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 18, 561–567 (1990).
Modrich, P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 281, 30305–30309 (2006).
Habano, W., Nakamura, S. & Sugai, T. Microsatellite instability in the mitochondrial DNA of colorectal carcinomas: evidence for mismatch repair systems in mitochondrial genome. Oncogene 17, 1931–1937 (1998).
Mason, P. A., Matheson, E. C., Hall, A. G. & Lightowlers, R. N. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 31, 1052–1058 (2003). The original report of mitochondrial MMR activity.
de Souza-Pinto, N. C. et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair 8, 704–719 (2009). This study was the first to identify a protein (YB1) involved in mitochondrial MMR.
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Hombauer, H., Campbell, C. S., Smith, C. E., Desai, A. & Kolodner, R. D. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell 147, 1040–1053 (2011).
Jürchott, K. et al. Identification of Y-box binding protein 1 as a core regulator of MEK/ERK pathway-dependent gene signatures in colorectal cancer cells. PLoS Genet. 6, e1001231 (2010).
Das, S. et al. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J. Biol. Chem. 282, 28474–28484 (2007).
Kadyrov, F. A. et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl Acad. Sci. USA 106, 8495–8500 (2009).
Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008).
Pavlov, Y. I., Mian, I. M. & Kunkel, T. A. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol. 13, 744–748 (2003).
Kadyrov, F. A., Dzantiev, L., Constantin, N. & Modrich, P. Endonucleolytic function of MutLα in human mismatch repair. Cell 126, 297–308 (2006).
Gaudreault, I., Guay, D. & Lebel, M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 32, 316–327 (2004).
Burkovics, P., Szukacsov, V., Unk, I. & Haracska, L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 34, 2508–2515 (2006).
Kim, M., Trinh, B. N., Long, T. I., Oghamian, S. & Laird, P. W. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 32, 5742–5749 (2004).
Tsuchimoto, D. et al. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 29, 2349–2360 (2001).
Kunkel, T. A. & Erie, D. A. DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 (2005).
Karran, P. & Bignami, M. DNA damage tolerance, mismatch repair and genome instability. Bioessays 16, 833–839 (1994).
Loughery, J. E. P. et al. DNMT1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Hum. Mol. Genet. 20, 3241–3255 (2011).
Robin, E. D. & Wong, R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell Physiol. 136, 507–513 (1988).
Shokolenko, I., Venediktova, N., Bochkareva, A., Wilson, G. L. & Alexeyev, M. F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 37, 2539–2548 (2009).
Mita, S., Monnat, R. J. Jr & Loeb, L. A. Resistance of HeLa cell mitochondrial DNA to mutagenesis by chemical carcinogens. Cancer Res. 48, 4578–4583 (1988).
Kasiviswanathan, R., Gustafson, M. A., Copeland, W. C. & Meyer, J. N. Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 287, 9222–9229 (2011).
Ikeda, S. & Ozaki, K. Action of mitochondrial endonuclease G on DNA damaged by L-ascorbic acid, peplomycin, and cis-diamminedichloroplatinum (II). Biochem. Biophys. Res. Commun. 235, 291–294 (1997).
Youle, R. J. & Narendra, D. P. Mechanisms of mitophagy. Nature Rev. Mol. Cell Biol. 12, 9–14 (2011).
Gao, Y. et al. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 471, 240–244 (2011). This and reference 3 demonstrate that the mitochondrial isoform of LIG3 is essential for cell viability.
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Santos, J. H., Meyer, J. N., Mandavilli, B. S. & Van Houten, B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314, 183–199 (2006).
Holt, I. J., Lorimer, H. E. & Jacobs, H. T. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100, 515–524 (2000).
Venø, S. T. et al. The human Suv3 helicase interacts with replication protein A and flap endonuclease 1 in the nucleus. Biochem. J. 440, 293–300 (2011).
Santos, J. H., Meyer, J. N., Skorvaga, M., Annab, L. A. & Van Houten, B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell 3, 399–411 (2004).
Minczuk, M., Lilpop, J., Boros, J. & Stepien, P. P. The 5′ region of the human hSUV3 gene encoding mitochondrial DNA and RNA helicase: promoter characterization and alternative pre-mRNA splicing. Biochim. Biophys. Acta 1729, 81–87 (2005).
Futami, K., Shimamoto, A. & Furuichi, Y. Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 30, 1685–1692 (2007).
Myers, K. A., Saffhill, R. & Oconnor, P. J. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis 9, 285–292 (1988).
Pohjoismäki, J. L. O. et al. Oxidative stress during mitochondrial biogenesis compromises mtDNA integrity in growing hearts and induces a global DNA repair response. Nucleic Acids Res. 40, 6595–6607 (2012).
Bell, O., Tiwari, V. K., Thomä, N. H. & Schübeler, D. Determinants and dynamics of genome accessibility. Nature Rev. Genet. 12, 554–564 (2011).
Lander, E. S. Initial impact of the sequencing of the human genome. Nature 470, 187–197 (2011).
Holt, I. J. et al. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion 7, 311–321 (2007).
Clayton, D. A., Doda, J. N. & Friedberg, E. C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc. Natl Acad. Sci. USA 71, 2777–2781 (1974).
Yasukawa, T. et al. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 25, 5358–5371 (2006).
Storici, F., Bebenek, K., Kunkel, T. A., Gordenin, D. A. & Resnick, M. A. RNA-templated DNA repair. Nature 447, 338–341 (2007).
Korhonen, J. A., Pham, X. H., Pellegrini, M. & Falkenberg, M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 23, 2423–2429 (2004).
Fuste, J. M. et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell 37, 67–78 (2010).
Reyes, A., Yang, M. Y., Bowmaker, M. & Holt, I. J. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 280, 3242–3250 (2005).
Jacobs, H. T. The mitochondrial theory of aging: dead or alive? Aging Cell 2, 11–17 (2003).
Gredilla, R., Garm, C., Holm, R., Bohr, V. A. & Stevnsner, T. Differential age-related changes in mitochondrial DNA repair activities in mouse brain regions. Neurobiol. Aging 31, 993–1002 (2010).
Imam, S. Z., Karahalil, B., Hogue, B. A., Souza-Pinto, N. C. & Bohr, V. A. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol. Aging 27, 1129–1136 (2006).
Chen, D. et al. Age-dependent decline of DNA repair activity for oxidative lesions in rat brain mitochondria. J. Neurochem. 81, 1273–1284 (2002).
Ahlqvist, K. J. et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in polg mutator mice. Cell Metab. 15, 100–109 (2012).
Hakonen, A. H. et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet. 77, 430–441 (2005).
Katyal, S. et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 26, 4720–4731 (2007).
Rouzier, C. et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy “plus” phenotype. Brain 135, 23–34 (2012).
Holt, I. J., Harding, A. E. & Morgan-Hughes, J. A. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 331, 717–719 (1988). The initial genetic evidence for mtDNA mutations resulting in human disease.
Krishnan, K. J. et al. What causes mitochondrial DNA deletions in human cells? Nature Genet. 40, 275–279 (2008).
Katada, H. & Komiyama, M. Artificial restriction DNA cutters to promote homologous recombination in human cells. Curr. Gene Ther. 11, 38–45 (2011).
Hua, S. B., Qiu, M., Chan, E., Zhu, L. & Luo, Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid 38, 91–96 (1997).
Sembongi, H., Di Re, M., Bokori-Brown, M. & Holt, I. J. The yeast Holliday junction resolvase, CCE1, can restore wild-type mitochondrial DNA to human cells carrying rearranged mitochondrial DNA. Hum. Mol. Genet. 16, 2306–2314 (2007).
Poulton, J. & Holt, I. J. Mitochondrial DNA: does more lead to less? Nature Genet. 8, 313–315 (1994).
Miller, F. J., Rosenfeldt, F. L., Zhang, C., Linnane, A. W. & Nagley, P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 31, e61 (2003).
Acknowledgements
We thank S. Wood, M. Minczuk, J. Pohjoismäki and J. Poulton for insights and discussions. L.K. is the recipient of a Cambridge University Commonwealth Trust Fellowship, and work in the authors' laboratory is supported by the UK Medical Research Council.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- N-glycosylic bond
-
The chemical bond between the base and the sugar of a nucleotide.
- Thymine glycol
-
5,6-dihydroxydihydrothymine induced by chemical oxidation or ionizing radiation (an oxidative mutagen).
- Apurinic/apyrimidinic site
-
(AP site). The site of an intact sugar-phosphate backbone after the nitrogenous base has been removed.
- β-elimination mechanism
-
Incision of the DNA backbone between the sugar and the phosphate (that is, 3′ to the sugar of the damaged nucleotide), yielding a 3′-phospho-α,β-unsaturated aldehyde (3′ PUA) and a 5′ phosphate on either side of the strand break.
- δ-elimination mechanism
-
Incision of the DNA backbone between the sugar and the phosphate (that is, 5′ to the sugar of the damaged nucleotide), removing the 3′ aldehyde from the β-elimination step to generate a 3′ phosphate adjacent to the 5′ phosphate.
- Nucleotide incision repair
-
A glycosylase-independent mechanism of DNA strand cleavage, 5′ to the damaged base, generating a 3′ hydroxyl that can be used for DNA synthesis, with the 5′ side of the strand break acting as a substrate for flap endonuclease 1 cleavage.
- Blocking residue
-
A 5′ phosphate and a 3′ hydroxyl are required for initiation of DNA synthesis and DNA ligation. A modification on either the 5′ or 3′ side of a strand break that does not support these processes is termed a blocking residue.
- Okazaki fragment maturation
-
The removal of primers during replication of the lagging strand to produce suitable substrates for DNA ligase to generate a continuous nascent strand of DNA.
- Nucleotide excision repair
-
A repair pathway that recognizes a diverse range of structural alterations that distort the shape of duplex DNA. After damage recognition, incisions are made on both sides of the lesion and a single-stranded DNA segment, containing the damage, is removed. The resulting DNA gap is subsequently filled in by DNA polymerase using the undamaged strand as a template.
- Insertion–deletion loops
-
(IDLs). Insertion or deletion of nucleotides on one strand of duplex DNA, which results in looping out from the linear structure of DNA.
- Microsatellites
-
Sequences of DNA that consist of repeating nucleotide units (1–6 base pairs in length). These can become longer or shorter owing to replication fork slippage, which can lead to frameshift mutations in genes, resulting in cancer predisposition.
- Translesion bypass
-
The continuity of replication (or transcription), despite the presence of damage on the template DNA, with a high likelihood of errors that may lead to mutation.
Rights and permissions
About this article
Cite this article
Kazak, L., Reyes, A. & Holt, I. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol 13, 659–671 (2012). https://doi.org/10.1038/nrm3439
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3439
This article is cited by
-
Mitochondrial E3 ligase MARCH5 is a safeguard against DNA-PKcs-mediated immune signaling in mitochondria-damaged cells
Cell Death & Disease (2023)
-
ATFS-1 counteracts mitochondrial DNA damage by promoting repair over transcription
Nature Cell Biology (2023)
-
Age-related mechanisms in the context of rheumatic disease
Nature Reviews Rheumatology (2022)
-
An HSP90 cochaperone Ids2 maintains the stability of mitochondrial DNA and ATP synthase
BMC Biology (2021)
-
DNA damage as a mechanism of neurodegeneration in ALS and a contributor to astrocyte toxicity
Cellular and Molecular Life Sciences (2021)