Abstract
Understanding the roles of splicing factors and splicing events during tumorigenesis would open new avenues for targeted therapies. Here we identify an oncofetal splicing factor, MBNL3, which promotes tumorigenesis and indicates poor prognosis of hepatocellular carcinoma patients. MBNL3 knockdown almost completely abolishes hepatocellular carcinoma tumorigenesis. Transcriptomic analysis revealed that MBNL3 induces lncRNA-PXN-AS1 exon 4 inclusion. The transcript lacking exon 4 binds to coding sequences of PXN mRNA, causes dissociation of translation elongation factors from PXN mRNA, and thereby inhibits PXN mRNA translation. In contrast, the transcript containing exon 4 preferentially binds to the 3′ untranslated region of PXN mRNA, protects PXN mRNA from microRNA-24–AGO2 complex-induced degradation, and thereby increases PXN expression. Through inducing exon 4 inclusion, MBNL3 upregulates PXN, which mediates the pro-tumorigenic roles of MBNL3. Collectively, these data demonstrate detailed mechanistic links between an oncofetal splicing factor, a splicing event and tumorigenesis, and establish splicing factors and splicing events as potential therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Torre, L. A. et al. Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 (2015).
Lee, J. S. et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 12, 410–416 (2006).
Becker, D. et al. Genetic signatures shared in embryonic liver development and liver cancer define prognostically relevant subgroups in HCC. Mol. Cancer 11, 55 (2012).
Yong, K. J. et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N. Engl. J. Med. 368, 2266–2276 (2013).
Bechara, E. G., Sebestyen, E., Bernardis, I., Eyras, E. & Valcarcel, J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52, 720–733 (2013).
Zhang, J. & Manley, J. L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 3, 1228–1237 (2013).
Oltean, S. & Bates, D. O. Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 (2014).
Kornblihtt, A. R. et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14, 153–165 (2013).
Lee, Y. & Rio, D. C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015).
Wang, Y. et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell 26, 374–389 (2014).
Zhou, X. et al. BCLAF1 and its splicing regulator SRSF10 regulate the tumorigenic potential of colon cancer cells. Nat. Commun. 5, 4581 (2014).
Salton, M. et al. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat. Commun. 6, 7103 (2015).
Lu, Z. X. et al. Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol. Cancer Res. 13, 305–318 (2015).
Holoch, D. & Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 (2015).
Fatica, A. & Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 (2014).
Yuan, J. H. et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014).
Wang, Y. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413–425 (2015).
McHugh, C. A. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015).
Wang, F. et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 60, 1278–1290 (2014).
Li, T. et al. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics 93, 235–242 (2009).
Lee, J. S. et al. Transcriptional ontogeny of the developing liver. BMC Genomics 13, 33 (2012).
Roessler, S. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 70, 10202–10212 (2010).
Chen, X. et al. Gene expression patterns in human liver cancers. Mol. Biol. Cell 13, 1929–1939 (2002).
Tung, E. K. et al. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 31, 1494–1504 (2011).
Zhu, P. et al. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J. Clin. Invest. 125, 3795–3808 (2015).
Lee, T. K. et al. CD24+ liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell 9, 50–63 (2011).
Shan, J. et al. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology 56, 1004–1014 (2012).
Han, H. et al. PBX3 is targeted by multiple miRNAs and is essential for liver tumour-initiating cells. Nat. Commun. 6, 8271 (2015).
Hoshida, Y. et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 144, 1024–1030 (2013).
Sznajder, L. J. et al. Mechanistic determinants of MBNL activity. Nucleic Acids Res. 44, 10326–10342 (2016).
Orengo, J. P., Bundman, D. & Cooper, T. A. A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic Acids Res. 34, e148 (2006).
Poulos, M. G. et al. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum. Mol. Genet. 22, 3547–3558 (2013).
Sen, A. et al. Paxillin mediates extranuclear and intranuclear signaling in prostate cancer proliferation. J. Clin. Invest. 122, 2469–2481 (2012).
Hu, C. T., Cheng, C. C., Wu, J. R., Pan, S. M. & Wu, W. S. PKCɛ-mediated c-Met endosomal processing directs fluctuant c-Met-JNK-paxillin signaling for tumor progression of HepG2. Cell Signal 27, 1544–1555 (2015).
Marasek, P. et al. Paxillin-dependent regulation of IGF2 and H19 gene cluster expression. J. Cell Sci. 128, 3106–3116 (2015).
Chen, D. L. et al. Overexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancer. Carcinogenesis 34, 803–811 (2013).
Kato, H. et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205, 1601–1610 (2008).
Gantier, M. P. & Williams, B. R. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 18, 363–371 (2007).
Wu, D. W., Chen, C. Y., Chu, C. L. & Lee, H. Paxillin confers resistance to tyrosine kinase inhibitors in EGFR-mutant lung cancers via modulating BIM and Mcl-1 protein stability. Oncogene 35, 621–630 (2016).
Jia, H. L. et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin. Cancer Res. 13, 1133–1139 (2007).
Kalsotra, A. et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA 105, 20333–20338 (2008).
Pascual, M., Vicente, M., Monferrer, L. & Artero, R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation 74, 65–80 (2006).
Wang, E. T. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150, 710–724 (2012).
Han, H. et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498, 241–245 (2013).
Kanadia, R. N. et al. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns 3, 459–462 (2003).
Zhao, W. et al. 1B50-1, a mAb raised against recurrent tumor cells, targets liver tumor-initiating cells by binding to the calcium channel α2delta1 subunit. Cancer Cell 23, 541–556 (2013).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730 (2008).
Yoon, J. H. et al. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655 (2012).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Wu, D. W. et al. Paxillin promotes tumor progression and predicts survival and relapse in oral cavity squamous cell carcinoma by microRNA-218 targeting. Carcinogenesis 35, 1823–1829 (2014).
Zhao, C. J., Du, S. K., Dang, X. B. & Gong, M. Expression of paxillin is correlated with clinical prognosis in colorectal cancer patients. Med. Sci. Monit. 21, 1989–1995 (2015).
Alisi, A. et al. Focal adhesion kinase (FAK) mediates the induction of pro-oncogenic and fibrogenic phenotypes in hepatitis C virus (HCV)-infected cells. PLoS ONE 7, e44147 (2012).
Wang, R. Y. et al. MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology 145, 1436–1448.e12 (2013).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Sharma, E., Sterne-Weiler, T., O’Hanlon, D. & Blencowe, B. J. Global mapping of human RNA–RNA interactions. Mol. Cell 62, 618–626 (2016).
El-Naggar, A. M. et al. Translational activation of HIF1α by YB-1 promotes sarcoma metastasis. Cancer Cell 27, 682–697 (2015).
Faller, W. J. et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500 (2015).
Acknowledgements
We thank T. A. Cooper from Baylor College of Medicine, USA, for the gift of the RG6 minigene. This work was supported by grants from the National Key Basic Research Program (973 project) (2015CB554004) from the Ministry of Science and Technology of China, the National Natural Science Foundation of China (Grant No. 81572792, 81330037, 81372240, and 81171937), the Shanghai Sailing Program (15YF1400100), and the Natural Science Foundation of Shanghai (15XD1504500 and 14JC1407800).
Author information
Authors and Affiliations
Contributions
S.-h.S., J.-h.Y. and F.W. designed the experiments. J.-h.Y., X.-n.L., T.-t.W. and W.P. performed the experiments. J.-h.Y., F.W., X.-n.L., T.-t.W., W.P. and Q.-f.T. analysed the data. W.-p.Z. and Q.-f.T. provided clinical specimens. J.-h.Y. wrote the manuscript. S.-h.S. supervised the project and helped with the writing of the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
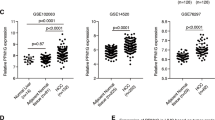
Supplementary Figure 1 MBNL3 expression is increased in HCC, and NANOG, OCT4, and SOX2 upregulate MBNL3 expression.
(a) Mbnl3 expression levels during mouse liver development from GSE13149 data. (b) Mbnl3 expression levels during mouse liver development from GSE21224 data. (c) MBNL3 expression levels in human HCC tissues (n = 268 independent samples) and noncancerous hepatic tissues (n = 243 independent samples) from GSE25097 data. P < 0.0001 by Mann–Whitney test. (d) MBNL3 expression levels in human HCC tissues (n = 247 independent samples) and noncancerous hepatic tissues (n = 239 independent samples) from GSE14520 data. P < 0.0001 by Mann–Whitney test. For c and d, the horizontal lines in the box plots represent the medians, the boxes represent the interquartile range, and the whiskers represent the minimum and maximum values. (e) The representative images of MBNL3 IHC staining in HCC tissues and noncancerous hepatic tissues. Scale bar, 50 μm. (f) Western blot analysis of MBNL3 expression in human immortalized, nontransformed liver cell line (QSG-7701), HCC cell lines (SMMC-7721, HCCLM3, MHCC97H, Huh7 and Hep3B), and hepatoblastoma cell line (HepG2). (g) Western blot analysis of NANOG, OCT4, and SOX2 expression in SMMC-7721 cells overexpressing NANOG, OCT4, and SOX2. (h) Western blot analysis of NANOG, OCT4, and SOX2 expression in NANOG, OCT4, and SOX2 depleted SMMC-7721 cells. (i) Western blot analysis of NANOG, OCT4, and SOX2 expression in human immortalized, nontransformed liver cell line (QSG-7701), HCC cell lines (SMMC-7721, HCCLM3, and Huh7), and hepatoblastoma cell line (HepG2). (j) Schematic outlining the predicted binding sites for NANOG, OCT4, and SOX2 in the MBNL3 promoter. (k) ChIP assays in SMMC-7721 cells using anti-NANOG, anti-OCT4, anti-SOX2, or nonspecific IgG. Enrichment was determined relative to input controls. Site N was used as negative control with no binding sites for NANOG, OCT4, and SOX2. Data are mean ± s.d. of n = 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. Source data are available in Supplementary Table 8. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 2 MBNL3 drives HCC tumorigenesis and is required for HCC tumorigenesis.
(ab) Tumors generated by subcutaneous injection with MBNL3 stably overexpressed SMMC-7721 cells into nude mice were stained for Ki67 (a) or TUNEL (b). n = 5 independent samples. Scale bars, 50 μm. (c,d) Tumors generated by subcutaneous injection with MBNL3 stably overexpressed QSG-7701 cells into nude mice were stained for Ki67 (c) or TUNEL (d). n = 4 independent samples. Scale bars, 50 μm. For a–d, data are mean ± s.d. ∗P < 0.05, ∗∗P < 0.01 by Mann–Whitney test. (e) Representative hematoxylin and eosin-stained images of liver tissues isolated from the mice at 28 day after intrasplenic injection with indicated SMMC-7721 cells. Scale bars, 500 μm. (f) The mRNA and protein levels of MBNL3 in MBNL3 stably silenced Huh7 cells. (g,h) Cell proliferations were measured using CCK-8 assays (g) or EdU immunofluorescence staining (h) in MBNL3 stably silenced Huh7 cells. Scale bars, 100 μm. (i) Colony formation assays of MBNL3 stably silenced Huh7 cells. Scale bars, 5 mm. (j) Cell apoptosis was detected by TUNEL staining in MBNL3 stably silenced Huh7 cells. Scale bars, 100 μm. For f–j, data are mean ± s.d. of n = 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. (k) Western blot analysis of apoptosis markers in MBNL3 stably silenced Huh7 cells. (l) Effects of MBNL3 knockdown in SMMC-7721 cells on subcutaneous tumor growth. Image of mice 28 days after injection is shown. (m) Effects of another independent shRNA knockdown of MBNL3 in SMMC-7721 cells on subcutaneous tumor growth. Image of mice 35 days after injection is shown. (n) Effects of MBNL3 knockdown in Huh7 cells on subcutaneous tumor growth. Images of mice and tumors 28 days after injection are shown. Scale bars, 1 cm. (o) Representative hematoxylin and eosin-stained images of liver tissues isolated from the mice 28 days after intrasplenic injection with indicated SMMC-7721 cells. Scale bars, 500 μm. Source data are available in Supplementary Table 8. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 3 Depletion of MBNL3 inhibits HCC cells growth in vitro and in vivo.
(a–d) The protein levels of MBNL3 in MBNL3 stably depleted MHCC97H (a), HCCLM3 (b), HepG2 (c), or Hep3B (d) cells. (e–h) Cell proliferations were measured using CCK-8 assays in MBNL3 stably depleted MHCC97H (e), HCCLM3 (f), HepG2 (g), or Hep3B (h) cells. (i–l) Cell proliferations were assessed using EdU immunofluorescence staining in MBNL3 stably depleted MHCC97H (i), HCCLM3 (j), HepG2 (k), or Hep3B (l) cells. Scale bars, 100 μm. (m-p) Cell apoptosis was detected by TUNEL staining in MBNL3 stably depleted MHCC97H (m), HCCLM3 (n), HepG2 (o), or Hep3B (p) cells. Scale bars, 100 μm. For e-p, data are mean ± s.d. of n = 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. (q–x) Effects of MBNL3 depletion on MHCC97H (q–u), HCCLM3 (r–v), HepG2 (s–w), or Hep3B (t–x) cells subcutaneous tumor growth. Tumor volumes were measured every 7 days (q–t). The mice were killed at 28 day after injection, and the tumors were excised and weighed (u–x). Data are mean ± s.d., n = 8 mice in each group for MHCC97H and HCCLM3 cells, n = 7 mice in each group for HepG2 and Hep3B cells, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Mann–Whitney test.
Supplementary Figure 4 MBNL3 induced alternative splicing of lncRNA-PXN-AS1.
(a) GSEA of gene signatures reported to be associated with poor or good survival in MBNL3 shRNA-2 stably transduced SMMC-7721 cells versus control cells. NES, normalized enrichment score. (b) Inclusion of exon 4 was examined in MBNL1 or MBNL2 overexpressed SMMC-7721 cells by RT-PCR. (c) LncRNA-PXN-AS1 isoforms expression levels was examined in SMMC-7721 cells overexpressing MBNL1 or MBNL2 by real-time qPCR. (d) A schematic showing PXN-AS1 minigene containing exon 4, 250bp of its upstream intron 3, and 302bp of its downstream intron 4. (e) The insert sequence of pre-lncRNA-PXN-AS1. The underlined sequences are exon 4. The previous reported putative MBNL3 binding motif YGCY were highlighted in red. (f–g) RT-PCR analysis of PXN-AS1 minigene coexpressed with MBNL3 expressing plasmid (f) or MBNL3 shRNA (g). (h) Binding of MBNL3 with pre-lncRNA-PXN-AS1 was detected by CLIP assays in SMMC-7721 cells exogenously expressing PXN-AS1 minigene with primer pairs complementary to MBNL3 intron 3, exon 4, intron 4, or S100A4 by real-time qPCR. (i) RT-PCR analysis of the effects of MBNL3 overexpression on wild type PXN-AS1 minigene (PXN-AS1-wt), PXN-AS1 minigene with mutation of the predicated MBNL3 binding sites in intron 3, exon 4, or intron 4. (j) Inclusion of exon 4 was examined in human fetal livers and adult livers by RT-PCR. (k) Inclusion of exon 4 was examined in human nontransformed liver cell line and HCC cell lines by RT-PCR. For b,c, and f–k, data are mean ± s.d. of n = 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01 by Student’s t-test. Source data are available in Supplementary Table 8. Uncropped images of gels are shown in Supplementary Fig. 9.
Supplementary Figure 5 Characterization of lncRNA-PXN-AS1 and the different effects of lncRNA-PXN-AS1 isoforms on PXN.
(a) Left; representative images of PCR products from the 5′-RACE or 3′-RACE procedure. The major PCR product is marked by an arrow. Right; sequencing of PCR products revealed the boundary of lncRNA-PXN-AS1 sequences. (b) The nucleotide sequence of full-length PXN-AS1-L and PXN-AS1-S. (c) The levels of PXN-AS1-L and PXN-AS1-S in purified polyadenylated RNAs. β-actin and U6 serve as poly (A) positive and negative control, respectively. (d) The levels of PXN-AS1-L and PXN-AS1-S in purified cytoplasmic or nuclear RNAs. β-actin and U6 serve as cytoplasmic and nuclear control, respectively. (e) Confocal RNA FISH images showed cytoplasmic localization of lncRNA-PXN-AS1. Scale bars, 10 μm. The probe detects both PXN-AS1-L and PXN-AS1-S. (f) PXN protein levels in PXN-AS1-L or PXN-AS1-S overexpressed QSG-7701 cells. (g) AMT crosslinking followed by proximity ligation of interacting RNA and sequencing confirmed the interactions between PXN-AS1-L exon 4 and PXN 3′UTR. (h) miR-24 expression in QSG-7701, SMMC-7721, HCCLM3, Huh7, and HepG2 cells. For c,d,h, data are mean ± s.d. of n = 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. (i) SMMC-7721 cell lysates were incubated with PXN full-length mRNA and biotin-labeled PXN-AS1-L, or PXN-AS1-L and biotin-labeled PXN full-length mRNA; after pull-down, bound proteins were extracted and assessed by western blot, and bound mRNAs were extracted and assessed by real-time qPCR. (j) Polysome profiles from SMMC-7721 cells incubated with harringtonine for 0 s (up) and 180 s (low) before harvest. The absorbance profile in 254 nm is outlined. (k,l) SMMC-7721 cells transiently expressing indicated transcripts of lncRNA-PXN-AS1 were incubated with harringtonine for 0 s and 180 s. Then polysomes in cytoplasmic extracts from these cells were fractionated through sucrose gradients, and the relative distributions of PXN (k) or β-actin (l) mRNAs in gradient fractions were measured by real-time qPCR, and polysomes fractions are shown as percentage of total RNA in the gradient. Source data are available in Supplementary Table 8. Uncropped images of gels and blots are shown in Supplementary Fig. 9.
Supplementary Figure 6 PXN modulates MCL1 and BIM expression, and promotes HCC tumorigenesis.
(a) PXN protein levels in SMMC-7721 cells stably overexpressing PXN. (b) Western blot analysis of MCL1 and BIM protein levels in SMMC-7721 cells stably overexpressing PXN. (c–f) Western blot analysis of MCL1 and BIM protein levels in SMMC-7721 (c) or QSG-7701(d) cells stably overexpressing MBNL3, SMMC-7721 (e) or Huh7 (f) cells stably silencing MBNL3. (g) Cell proliferations were measured using CCK-8 assays in SMMC-7721 cells stably overexpressing PXN. (h) Cell proliferations were assessed using EdU immunofluorescence staining in SMMC-7721 cells stably overexpressing PXN. Scale bars, 100 μm. (i) Colony formation assays of SMMC-7721 cells stably overexpressing PXN. Scale bars, 5 mm. (j) Cell apoptosis was detected by TUNEL staining in SMMC-7721 cells stably overexpressing PXN. Scale bars, 100 μm. For g,j, data are mean ± s.d. of n = 3 independent experiments ∗P < 0.05, ∗∗P < 0.01 by Student’s t-test. (k) Western blot analysis of apoptosis markers in SMMC-7721 cells stably overexpressing PXN. (l,n) Effects of PXN overexpression in SMMC-7721 cells on subcutaneous tumor growth. Tumor volumes were measured every 7 days (l). The mice were killed 21 days after injection, and the tumors were excised and weighed (m,n). Scale bars, 1 cm. Data are mean ± s.d. of n = 10 mice in each group, ∗∗P < 0.01 by Mann–Whitney test. Source data are available in Supplementary Table 8. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 7 The biologic effects of MBNL3 on HCC cells tumorigenesis are partially medicated by AS of lncRNA-PXN-AS1.
(a) SMMC-7721 cells with stable coexpression of shMBNL3 and PXN-AS1-L were generated. PXN-AS1-L expression levels in indicated cells were confirmed by real-time qPCR. (b) PXN protein levels in indicated SMMC-7721 cells. (c,d) Cell proliferations were measured using CCK-8 assays (c) or EdU immunofluorescence staining (d) in indicated cells. Scale bars, 100 μm. (e) Cell apoptosis was detected by TUNEL staining in indicated cells. Scale bars, 100 μm. (f) Western blot analysis of apoptosis markers in indicated cells. (g,h) PXN-AS1-L partially rescues the effects of MBNL3 depletion on subcutaneous tumor growth. Tumor volumes were measured every 7 days (g). The mice were killed 28 days after injection, and the tumors were excised and weighed (h). Scale bars, 1 cm. (i) SMMC-7721 cells with stable coexpression of MBNL3 and PXN-AS1-S were generated. PXN-AS1-S expression levels in indicated cells were confirmed by real-time qPCR analysis. (j) PXN protein levels in indicated SMMC-7721 cells. (k,l) Cell proliferations were measured using CCK-8 assays (k) or EdU immunofluorescence staining (l) in indicated cells. Scale bars, 100 μm. (m) Cell apoptosis was detected by TUNEL staining in indicated cells. Scale bars, 100 μm. (n) Western blot analysis of apoptosis markers in indicated cells. (o,p) PXN-AS1-S partially rescues the effects of MBNL3 overexpression on subcutaneous tumor growth. Tumor volumes were measured every 7 days (o). The mice were killed 28 days after injection, and the tumors were excised and weighed (p). Scale bars, 1 cm. For a,c–e,i and k-m data are mean ± s.d. of n = 3 independent experiments ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. For g,h,o,p, data are mean ± s.d. of n = 8 mice in each group for g,h, and n = 7 mice in each group for o,p ∗P < 0.05, ∗∗P < 0.01,∗∗∗P < 0.001 by Mann–Whitney test. Source data are available in Supplementary Table 8. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary Figure 8 Depletion of PXN abolishes the biological effects of MBNL3 overexpression.
(a) SMMC-7721 cells with stable coexpression of MBNL3/shPXN or controls were generated. The protein levels of PXN in indicated cells were confirmed by western blot. (b) Cell proliferations were measured using CCK-8 assays in indicated SMMC-7721 cells. (c) Cell proliferations were assessed using EdU immunofluorescence staining in indicated SMMC-7721 cells. Scale bars, 100 μm. (d) Colony formation assays of indicated SMMC-7721 cells. Scale bars, 5 mm. (e) Cell apoptosis was detected by TUNEL staining in indicated SMMC-7721 cells. Scale bars, 100 μm. For b–e, data are mean ± s.d. of n = 3 independent experiments ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 by Student’s t-test. (f) Western blot analysis of apoptosis markers in indicated SMMC-7721 cells. (g–i) Depletion of PXN abolishes the effects of MBNL3 overexpression on subcutaneous tumor growth. Tumor volumes were measured every 7 days (g). The mice were killed at 21 day after injection, and the tumors were excised and weighed (h–i). Scale bars, 1 cm. Data are mean ± s.d. of n = 8 mice in each group ∗∗∗P < 0.001 by Mann–Whitney test. Source data are available in Supplementary Table 8. Uncropped images of blots are shown in Supplementary Fig. 9.
Supplementary information
Supplementary Information
Supplementary Information (PDF 5797 kb)
Supplementary Table 1
Supplementary Information (XLSX 10 kb)
Supplementary Table 2
Supplementary Information (XLSX 9 kb)
Supplementary Table 3
Supplementary Information (XLSX 10 kb)
Supplementary Table 4
Supplementary Information (XLSX 16 kb)
Supplementary Table 5
Supplementary Information (XLSX 56 kb)
Supplementary Table 6
Supplementary Information (XLSX 10 kb)
Supplementary Table 7
Supplementary Information (XLSX 12 kb)
Supplementary Table 8
Supplementary Information (XLSX 100 kb)
Rights and permissions
About this article
Cite this article
Yuan, Jh., Liu, Xn., Wang, Tt. et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol 19, 820–832 (2017). https://doi.org/10.1038/ncb3538
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3538
This article is cited by
-
Elevated expression of WSB2 degrades p53 and activates the IGFBP3-AKT-mTOR-dependent pathway to drive hepatocellular carcinoma
Experimental & Molecular Medicine (2024)
-
Role of UPF1 in lncRNA-HEIH regulation for hepatocellular carcinoma therapy
Experimental & Molecular Medicine (2024)
-
METTL3 and METTL14-mediated N6-methyladenosine modification of SREBF2-AS1 facilitates hepatocellular carcinoma progression and sorafenib resistance through DNA demethylation of SREBF2
Scientific Reports (2024)
-
A Novel lncRNA lncRNA-4045 Promotes the Progression of Hepatocellular Carcinoma by Affecting the Expression of AKR1B10
Digestive Diseases and Sciences (2024)
-
Breast metastatic tumors in lung can be substituted by lung-derived malignant cells transformed by alternative splicing H19 lncRNA
Breast Cancer Research (2023)