Abstract
Cancer stem cells (CSCs) may be responsible for tumour dormancy, relapse and the eventual death of most cancer patients1. In addition, these cells are usually resistant to cytotoxic conditions. However, very little is known about the biology behind this resistance to therapeutics. Here we investigated stem-cell death in the digestive system of adult Drosophila melanogaster. We found that knockdown of the coat protein complex I (COPI)–Arf79F (also known as Arf1) complex selectively killed normal and transformed stem cells through necrosis, by attenuating the lipolysis pathway, but spared differentiated cells. The dying stem cells were engulfed by neighbouring differentiated cells through a draper–myoblast city–Rac1–basket (also known as JNK)-dependent autophagy pathway. Furthermore, Arf1 inhibitors reduced CSCs in human cancer cell lines. Thus, normal or cancer stem cells may rely primarily on lipid reserves for energy, in such a way that blocking lipolysis starves them to death. This finding may lead to new therapies that could help to eliminate CSCs in human cancers.
Similar content being viewed by others
Main
To investigate the molecular mechanism behind the resistance of CSCs to therapeutics, we studied the death of stem cells with different degrees of quiescence in the adult Drosophila digestive system, including intestinal stem cells (ISCs)2,3, renal and nephric stem cells (RNSCs)4 and hindgut intestinal stem cells (HISCs)5,6 (Fig. 1a and Extended Data Fig. 1a). We found that expression of the proapoptotic genes rpr and p53 effectively ablated differentiated cells but had little effect on stem cells (Extended Data Fig. 1b–n).
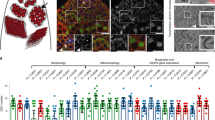
a, Diagram of three types of stem cells near the hindgut–midgut junction, and the cells in which esg–Gal4 (esg) and wg–Gal4 (wg) were expressed. b–i, Representative images of the posterior midguts of flies with the indicated phenotypes. b, esgts > upd, 29 °C, 4 d (n = 33). c, esgts > upd + rpr, 29 °C, 4 d (n = 35). d, esgts > RasV12, 29 °C, 7 d (n = 34). e, esgts > RasV12 + rpr, 29°C, 7 d (n = 29). f, esgts > RasV12 + Arf79FRNAi, 29 °C, 7 d (n = 35). g, esgts > NDN, 29 °C, 7 d (n = 24). h, esgts > NDN + rpr, 29 °C, 7 d (n = 29). i, esgts > NDN + Arf79FRNAi, 29 °C, 7 d (n = 38). j, Quantification of GFP+ cells from midguts isolated from flies with the indicated genotypes. Data are represented as mean ± s.e.m. Statistical significance determined by Student’s t-test, ***P < 0.0001. The posterior midguts of flies with the indicated genotypes were dissected, stained with the GFP and Prospero (Pros) antibodies and analysed by confocal microscopy. White arrows in b and c point to the hindgut–midgut junction. g, h, Red arrows with white dotted lines point to clusters of ISCs enteroblasts and yellow arrows with yellow dotted lines point to clusters of enteroendocrine cells. i, Red and yellow arrows point to remaining ISCs/enteroblasts, and enteroendocrine cells, respectively. Scale bars in b–i, 10 μm.
In mammals, treatment-resistant leukaemic stem cells (LSCs) can be eliminated by a two-step protocol involving initial activation by interferon-α (IFNα) or colony-stimulating factor (G-CSF), followed by targeted chemotherapy7. In Drosophila, activation of the hopscotch (also known as JAK)–Stat92E signalling pathway induces hyperplastic stem cells, which are overproliferating, but retain their apico-basal polarity and differentiation ability4,6,8. We conducted a slightly different two-step protocol in Drosophila stem cells by overexpressing the JAK–Stat92E pathway ligand unpaired (upd) and rpr together. The induction of upd + rpr using the temperature-sensitive (ts) mutant esg–Gal4 (esgts > upd + rpr; Fig. 1b, c, j and Extended Data Fig. 1o–q) effectively ablated all of the ISCs and RNSCs through apoptosis within four days. Consistent with this result, expressing a gain-of-function Raf mutant (Rafgof) also accelerated apoptotic cell death of hyperplastic ISCs9.
Expressing a constitutively active form of Ras oncogene at 85D (also known as RasV12) in RNSCs10 and the knockdown of Notch activity in ISCs11,12 can transform these cell types into CSC-like neoplastic stem cells, which were not only overproliferating, but also lost their apico-basal polarity and differentiation ability (Fig. 1d, g). We found that expressing rpr in RasV12-transformed RNSCs (esgts > RasV12 + rpr; Fig. 1e, j) or in ISCs expressing a dominant–negative form of Notch (NDN) (esgts > NDN + rpr; Fig. 1h, j) caused the ablation of only a proportion of the transformed RNSCs and few transformed ISCs and it did not affect differentiated cells (Extended Data Fig. 1r–u); substantial populations of the neoplastic stem cells remained even seven days after rpr induction.
These results suggest that the activation of proliferation can accelerate the apoptotic cell death of hyperplastic stem cells, but that a proportion of actively proliferating neoplastic RNSCs and ISCs are resistant to apoptotic cell death. Neoplastic tumours in Drosophila are more similar to high-grade malignant human tumours than are the hyperplastic Drosophila tumours13.
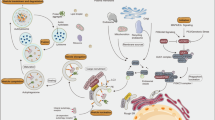
Vesicle-mediated COPI and COPII are essential components of the trafficking machinery for vesicle transportation between the endoplasmic reticulum and the Golgi14. In addition, the COPI complex regulates the transport of lipolysis enzymes to the surface of lipid droplets for lipid droplet usage15 (Extended Data Fig. 2a). In our previous screen, we found that knockdown of COPI components (including Arf79F, the Drosophila homologue of ADP-ribosylation factor 1 (Arf1)) rather than COPII components16 resulted in stem-cell death, suggesting that lipid-droplet usage (lipolysis) rather than the general trafficking machinery between the endoplasmic reticulum and Golgi is important for stem-cell survival.
To further investigate the roles of these genes in stem cells, we used a recombined double Gal4 line of esg–Gal4 and wg–Gal4 to express genes in ISCs, RNSCs, and HISCs (esgts wgts > X). Knockdown of these genes using RNA interference (RNAi) in stem cells (esgts wgts > XRNAi; Extended Data Fig. 2b–k) ablated most of the stem cells in 1 week. However, expressing Arf79FRNAi in enterocytes (NP1ts > Arf79FRNAi; Extended Data Fig. 2l–o) or in differentiated stellate cells in Malpighian tubules (tshts > Arf79FRNAi; Extended Data Fig. 2p, q) did not cause similar marked ablation. These results suggest that Arf79F knockdown selectively kills stem cells and not differentiated cells.
We also found that expressing Arf79FRNAi (esgts > RasV12 + Arf79FRNAi; Fig. 1f, j) or ζ-COPRNAi (esgts wgts >RasV12 + ζ-COPRNAi; Extended Data Fig. 2r) in RasV12-transformed RNSCs ablated almost all of the transformed stem cells. Similarly, expressing Arf79FRNAi (esgts > NDN + Arf79FRNAi; Fig. 1i, j) or δ-COPRNAi (esgts wgts > NDN + δ-COPRNAi; Extended Data Fig. 2s) in NDN-transformed ISCs ablated all of the cells within one week, but restored differentiated cells to close to their normal levels within one week (Extended Data Fig. 2t, u).
We further generated δ-COP- and γ-COP-mutant clones using the mosaic analysis with a repressible cell marker (MARCM) technique17 and found that the COPI complex cell-autonomously regulated stem cell survival (Extended Data Fig. 3a–h). In summary, knockdown of the COPI–Arf79F complex effectively ablated normal and transformed stem cells but not differentiated enterocytes or stellate cells.
In our RNAi screen we also identified acyl-CoA synthetase long-chain (ACSL), an enzyme in the Drosophila lipolysis–β-oxidation pathway16,18 (Extended Data Fig. 2a), and bubblegum (bgm), a very long-chain fatty acid-CoA ligase16,19. RNAi-mediated knockdown of Acsl (esgts wgts > AcslRNAi; Extended Data Fig. 2i, k) and bgm (esgts wgts > bgmRNAi; Extended Data Fig. 2j, k) effectively killed ISCs and RNSCs, but killed HISCs less effectively. Expressing AcslRNAi in RasV12-transformed RNSCs (esgts wgts > RasV12 + AcslRNAi; Extended Data Fig. 2v) also ablated almost all of the transformed RNSCs in one week.
Brummer (bmm) is a triglyceride lipase, the Drosophila homologue of mammalian ATGL, the first enzyme in the lipolysis pathway20 (Extended Data Fig. 3a). Scully (scu) is the Drosophila orthologue of hydroxy-acyl-CoA dehydrogenase, an enzyme in the β-oxidation pathway21. Hepatocyte nuclear factor 4 (Hnf4) regulates the expression of several genes involved in lipid mobilization and β-oxidation21. To determine whether the lipolysis–β-oxidation pathway is required for COPI–Arf79F-mediated stem cell survival, we expressed upstream activating sequence (UAS)-regulated constructs (UAS–bmm (Fig. 2c, h), UAS–Hnf4 (Fig. 2d, h), and UAS–scu (Fig. 2e, h)) in stem cells that were depleted of Arf79F (Fig. 2b–e), β-COP (Extended Data Fig. 2w), or ζ-COP (Extended Data Fig. 2x). Overexpressing either scu or Hnf4 significantly (P < 0.0001) attenuated the stem cell death caused by knockdown of the COPI–Arf79F complex. Expressing UAS–Hnf4 (Fig. 2i) and UAS–scu (Fig. 2j) in FRT82B–γ-COP10 MARCM clones also rescued the stem cell death phenotype induced by γ-COP knockdown (Extended Data Fig. 3f, h). However, bmm overexpression did not rescue the stem-cell death induced by Arf79F knockdown (Fig. 2c, h). Since there are several other triglyceride lipases in Drosophila in addition to bmm, another lipase may redundantly regulate the lipolysis pathway.
a–g, Representative images are shown. The genotypes of the flies in each panel were: a, esgts wgts > lacZRNAi, 29 °C, 7 d (n = 34). b, esgts wgts > Arf79FRNAi, 29 °C, 7 d (n = 38). c, esgts wgts > Arf79FRNAi + bmm, 29 °C, 7 d (n = 31). d, esgts wgts > Arf79FRNAi + Hnf4, 29 °C, 7 d (n = 29). e, esgts wgts > Arf79FRNAi + scu, 29 °C, 7 d (n = 32). f, esgts wgts > Arf79FRNAi + p35, 29 °C, 7 d (n = 37). g, esgts wgts > Arf79FRNAi + Atg12RNAi, 29 °C, 7 d (n = 32). h, Quantification of GFP+ cells from midguts isolated from flies with the indicated genotypes. Data are represented as mean ± s.d. Statistical significance determined by Student’s t-test, ***P < 0.0001. NS, not significant (P > 0.05). i–k, MARCM clones of flies with the following genotypes: i, UAS–Hnf4; FRT82B–γ-Cop10, 7 d (n = 30) after clonal induction (ACI). j, UAS-scu; FRT82B-γ-Cop10, 7 d ACI (n = 32). k, UAS–cat; FRT82B–γ-Cop10, 7 d ACI (n = 35). The posterior midguts of flies with the indicated genotypes were dissected, stained with the GFP, Prospero (Pros) and Armadillo (Arm) antibodies and analysed by confocal microscopy. White arrows in a–g point to the hindgut–midgut junction. White dotted lines in i–k outline GFP+ clones. Scale bars in a–g and i–k, 10 μm.
To further investigate the function of lipolysis in stem cells, we investigated the expression of a lipolysis reporter (GAL4–dHFN4; UAS–nlacZ)21, which consisted of hsp70–GAL4–dHNF4 combined with a UAS–nlacZ reporter gene. The flies were either cultured continuously at 29 °C or heat-shocked for 30 min at 37 °C, 12 h before dissection. Without heat shock, the reporter was expressed only in ISCs and RNSCs of mature adult flies, but not in enteroendocrine cells, enterocytes, quiescent HISCs or quiescent ISCs of freshly emerged young adult flies (less than 3 days old) (Extended Data Fig. 3i–m). Expressing δ-COPRNAi (esgts > δ-COPRNAi + GAL4–dHFN4; UAS–nlacZ) almost completely eliminated the reporter expression (Extended Data Fig. 3n), suggesting that the reporter was specifically regulated by the COPI complex. After heat shock or when a constitutively active form of JAK (hopTum-l) was expressed, the reporter was strongly expressed in ISCs, RNSCs and HISCs, but not in enteroendocrine cells or enterocytes (Extended Data Fig. 3o, p). These data suggest that COPI-complex-regulated lipolysis was active in stem cells, but not in differentiated cells, and that the absence of the reporter expression in quiescent HISCs at 29 °C was probably owing to weak hsp70 promoter activity rather than to low lipolysis in these cells.
We further investigated lipid storage, and found that the size and number of lipid droplets were markedly increased in stem cells after knockdown of Arf79F (esgts > Arf79FRNAi) (Extended Data Fig. 3q–v).
We also used Arf1 inhibitors (brefeldin A, golgicide A, secin H3, LM11 and LG8) and fatty-acid-oxidation (FAO) inhibitors (triacsin C, mildronate, etomoxir and enoximone) and found that these inhibitors markedly reduced stem-cell tumours in Drosophila through the lipolysis pathway but had a negligible effect on normal stem cells (Extended Data Fig. 4).
These data together suggest that the COPI–Arf1 complex regulates stem-cell survival through the lipolysis–β-oxidation pathway, and that knockdown of these genes blocks lipolysis but promotes lipid storage. Further, the transformed stem cells are more sensitive to Arf1 inhibitors and may be selectively eliminated by controlling the concentration of Arf1 inhibitors.
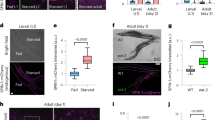
Our data suggest that neither caspase-mediated apoptosis nor autophagy-regulated cell death regulates the stem-cell death induced by the knockdown of components of the COPI–Arf79F complex (Fig. 2f–h). We therefore investigated whether necrosis regulates the stem-cell death induced by knockdown of the COPI–Arf79F complex. Necrosis is characterized by early plasma membrane rupture, reactive oxygen species (ROS) accumulation and intracellular acidification22. Propidium iodide detects necrotic cells with compromised membrane integrity, the oxidant-sensitive dye dihydroethidium (DHE) indicates cellular ROS levels and LysoTracker staining detects intracellular acidification22. We detected the membrane rupture phenotype only in esgts.wgts > Arf79FRNAi ISCs but not in wild-type ISCs (Figs 3a–d, 4a and Extended Data Fig. 5a–i) and the propidium iodide signal was observed only in ISCs from flies that had RNAi-induced knockdown of expression of COPI–Arf79F components (esgts wgts > XRNAi, Fig. 3g–i, l and Extended Data Fig. 5k–p), and not in cells from wild-type (Fig. 3e, f, l and Extended Data Fig. 5j, p), scu-rescued Arf79FRNAi (esgts wgts > scu + Arf79FRNAi, Fig. 3j, l) or Hnf4-rescued Arf79FRNAi (esgts wgts > Hnf4 + Arf79FRNAi, Fig. 3k, l) flies. In the esgts wgts > AcslRNAi flies, all of the ISCs and RNSCs were ablated after four days at 29 °C, but a fraction of the HISCs remained, and these were also propidium iodide positive (Extended data Fig. 5n–p), indicating that the HISCs were dying slowly. This slowness may have been due to either a lower GAL4 (wg–Gal4) activity in these cells compared to ISCs and RNSCs (esg–Gal4) or quiescence of the HISCs. Furthermore, strong propidium iodide signals were detected in transformed ISCs from esgts > NDN + Arf79FRNAi but not esgts > NDN flies (Extended data Fig. 6a–d), indicating that the transformed stem cells were dying through necrosis.
The genotypes of the flies in each panel were: a, b, e, f, esgts wgts > lacZRNAi, 29 °C, 4 d (n = 32). c, d, g, h, esgts wgts > Arf79FRNAi, 29 °C, 4 d (n = 28). i, esgts wgts > δ-CopRNAi, 29 °C, 4 d (n = 30). j, esgts wgts > Arf79FRNAi + scu, 29 °C, 4 d (n = 28). k, esgts wgts > Arf79FRNAi + Hnf4, 29 °C, 4 d (n = 30). l, Quantification of PI+ cells from midguts isolated from flies with the indicated genotypes. Data are represented as mean ± s.d. Statistical significance determined by Student’s t-test, ***P < 0.0001; NS, not significant (P > 0.05). The posterior midguts of flies with the indicated genotypes were dissected, stained with GFP and Armadillo (Arm) antibodies or indicated reagents, and analysed by confocal microscopy. White arrows in a–i point to esg–GFP+ cells and in j–k point to the hindgut–midgut junction. White dotted lines in d outline ISCs/enteroblasts, yellow dotted lines outline enterocytes. Scale bars in a–k, 10 μm.
a, Representative images from esgts > Arf79FRNAi cells, 29°C, 7 d (n = 32). A dying ISC is engulfed by a neighbouring enterocyte. The posterior midguts of flies were dissected, stained with the GFP and Armadillo (Arm) antibodies, and analysed by confocal microscopy. Arrow points to GFP+ stem cell/progenitors and the dotted line outlines a nucleus and cell membrane of an enterocyte. Scale bar, 10 μm. b, Quantification of Dl+ (Delta+) ISCs cells in flies with the NP1ts esgts driver and indicated genotypes (See images in Extended Data Fig. 9 for details). Data are represented as mean ± s.e.m. Statistical significance determined by Student’s t-test, *P < 0.05, ***P < 0.0001. NS, not significant (P > 0.05). c, Model of ISC death induced by knockdown of the COPI–Arf1 complex. Details are described in the text. The autophagosome is involved in the last step of phagocytosis (degradation of internalized cargo)27, and autophagy may both function downstream of and be regulated by the drpr–Mbc–Rac1–JNK pathway.
Similarly, DHE (Extended Data Fig. 6e–h) or LysoTracker (Extended Data Fig. 6i–l) signals were detected only in ISCs from esgts > Arf79FRNAi flies (Extended Data Fig. 6g, h, k, l), but not from wild-type flies (Extended Data Fig. 6e, f, i, j), indicating that the dying ISCs had accumulated ROS and were intracellularly acidified. Overexpressing catalase (a ROS-chelating enzyme) rescued the stem-cell death specifically induced by the γ-COP mutant clone (Fig. 2k) or by Arf79F knockdown (Extended Data Fig. 7b), and the ROS inhibitor NAC blocked the Arf1 inhibitor-induced death of RasV12-induced RNSC tumours (Extended Data Fig. 4i, l). These data together suggest that knockdown of the COPI–Arf1 complex induced the death of stem cells or of transformed stem cells (RasV12–RNSCs, NDN–ISCs) through ROS-induced necrosis. Although ISCs, RNSCs, and HISCs exhibit different degrees of quiescence, they all rely on lipolysis for survival, suggesting that this is a general property of stem cells.
We noticed cases where the GFP-positive material of the dying ISCs was present within neighbouring enterocytes (Fig. 4a, Extended Data Fig. 5a–i), suggesting that these enterocytes had engulfed dying ISCs.
The JNK pathway, autophagy and engulfment genes are involved in the engulfment of dying cells23,24. We therefore investigated whether these genes are required for COPI–Arf79F-regulated ISC death. We found that: (1) ISC death activated JNK signalling and autophagy in neighbouring enterocytes (Extended Data Fig. 7i–n); (2) knockdown of these genes in enterocytes but not in ISCs rescued ISC death to different degrees (Fig. 4b and Extended Data Figs 8a–i, 9a–l); (3) the drpr–mbc–Rac1–JNK pathway in enterocytes is not only necessary but also sufficient for ISC death (Extended Data Figs 8j–n and 9m, n); and (4) inhibitors of JNK and Rac1 could block Arf1-inhibitor-induced cell death of the RasV12-induced RNSC tumours (Extended Data Fig. 4g, h, l). These data together suggest that the drpr–mbc–Rac1–JNK pathway in neighbouring differentiated cells controls the engulfment of dying or transformed stem cells (Fig. 4c).
Our finding that the COPI–Arf79F–lipolysis–β-oxidation pathway regulated transformed stem-cell survival in the fly led us to investigate whether the pathway has a similar role in CSCs. We tested two Arf1 inhibitors (brefeldin A and golgicide A) and two FAO inhibitors (triascin C and etomoxir) on human cancer cell lines, and found that the growth, tumoursphere formation and expression of tumour-initiating cell markers of the four cancer cell lines were significantly (P < 0.01) suppressed by these inhibitors (Extended Data Fig. 10), suggesting that these inhibitors suppress CSCs. In mouse xenografts of BSY-1 human breast cancer cells, a novel low-cytotoxicity Arf1-ArfGEF inhibitor called AMF-26 was reported to induce complete regression in vivo in five days25. Together, this report and our results suggest that inhibiting Arf1 activity or blocking the lipolysis pathway can kill CSCs and block tumour growth.
Stem cells or CSCs are usually localized to a hypoxic storage niche, surrounded by a dense extracellular matrix26, which may make them less accessible to sugar and amino acid nutrition from the body’s circulatory system. Most normal cells rely on sugar and amino acids for their energy supply, with lipolysis playing only a minor role in their survival. Our results suggest that stem cells and CSCs are metabolically unique; they rely mainly on lipid reserves for their energy supply, and blocking COPI–Arf1-mediated lipolysis can starve them to death. We further found that transformed stem cells were more sensitive than normal stem cells to Arf1 inhibitors (Extended Data Fig. 4). Thus, selectively blocking lipolysis may kill CSCs without severe side effects. Therefore, targeting the COPI–Arf1 complex or the lipolysis pathway may prove to be a well-tolerated, novel approach for eliminating CSCs.
Methods
Fly strains
The following fly strains were used: NP1–Gal4 and FRT19A–δ-COPG0051 (from DGRC); esg–Gal4 and tsh–Gal4 (from S. Hayashi); wg–Gal4 (from J.-P. Vincent); UAS–upd (generated in our laboratory); UAS–NDN(2X) (from M. Fortini); UAS–bmm (from R. P. Kuhnlein); UAS–Hnf4 and GAL4–dHFN4 ; UAS–nlacZ (from C. Thummel); mira–GFP (from F. Schweisguth); pmCherry–Atg8a (from E. Baehrecke); UAS–GAP43–mCherry (from T. Lecuit); UAS–drprRNAi and UAS–drpr (from M. Freeman); UAS–DJunAsp (from M. Mlodzik); UAS–hepCA, UAS–RasV12, UAS–scu, UAS–rpr, UAS–p53, UAS–p35, UAS–Cat, UAS–Sod, UAS–Sod2, UAS–RacDN(N17), UAS–RacV12, UAS–bskDN, hopTum-l, puc–lacZ (pucE69), UAS–2XEYFP, tub–Gal80ts, and fly lines used for MARCM clones (FRT19A, tub–Gal80; FRT82B, tub–Gal80; SM6, hs–flp; MKRS, hs–flp; act>y+>Gal4, UAS–GFP; FRT82B–γ-COP10; and UAS–γ-COP FRT82B–γ-COP10) were obtained from the Bloomington Drosophila Stock Center at Indiana University.
RNAi stocks
An upstream activating sequence (UAS)-regulated double-stranded inverse-repeat construct was designed to target Arf79F: (UAS–Arf79FRNAi)-VDRC Transformant ID: 23082 (v23082). The RNA level was reduced to 39.0% in the Act–Gal4–UAS–Arf79FRNAi flies (ref. 16), and the phenotypes were confirmed by two independent RNAi lines (v103572 and v23080). The other RNAi lines used were: δ-COP–v41551; RNA level was reduced to 14.3% in Act–Gal4–UAS–δ-COPRNAi flies (ref. 16), and phenotypes were confirmed by an independent RNAi line (Bloomington stock number 31764 (BL31764 (TRiP ID HM04076)). β-COP–BL31709 (TRiP ID HM04016); RNA level was reduced to 13.3% in Act–Gal4–UAS–β-COPRNAi flies (ref. 16), and phenotypes were confirmed by two independent RNAi lines (v109641 and v15418). β’-COP–BL31710 (TRiP ID HM04017); RNA level was reduced to 3.2% in Act–Gal4–UAS–β’-COPRNAi flies (ref. 16). ζ-COP-BL28960 (TRiP ID HM05171); RNA level was reduced to 47.0% in Act–Gal4–UAS–ζ-COPRNAi flies (ref. 16), and phenotypes were confirmed by two independent RNAi lines (v34768 and v104405). garz-BL31232 (TRiP ID JF01013); RNA level was reduced to 52.4% in the Act–Gal4–UAS–garzRNAi flies (ref. 16). Acsl–BL27729 (TRiP ID JF02811); RNA level was reduced to 25.5% in Act–Gal4–UAS–AcslRNAi flies (ref. 16). bgm-v34854; RNA level was reduced to 56.2% in Act–Gal4–UAS–bgmRNAi flies (ref. 16), and phenotypes were confirmed by two independent RNAi lines (v105635 and BL28639 (TRiP ID JF03054)). γ-COP–BL28889 (TRiP IDHM05099); Atg5–BL34899 (TRiP ID HMS01244); Atg12RNAi (from E. Baehrecke, ref. 28); mbc–BL32355 (TRiP ID HMS00346); PSR–BL33700 (TRiP ID HMS00576); mys–BL33642 (TRiP ID HMS00043); CycT–BL32976 (TRiP ID HMS00776). The sequences used for each VDRC knock-down strain are available at https://stockcenter.vdrc.at) and for each Bloomington knock-down strain at http://flystocks.bio.indiana.edu. The data presented in all figures were generated by using the first RNAi line for all genes.
MARCM clone assay
To induce MARCM clones, three- or four-day-old adult female flies were heat-shocked twice with an interval of 8–12 h at 37 °C for 60 min. The flies were transferred to fresh food daily after the final heat shock and their posterior midguts were processed for staining at the indicated times.
RNAi-mediated gene depletion
To target the expression of UAS-linked genes in the cell types of interest, we used specific drivers. The posterior midgut of adult Drosophila is maintained by multipotent ISCs2,3, which differentiate into secretory enteroendocrine cells and absorptive enterocytes through immature enteroblasts. Enterocytes are polyploid and express the transcription factor Pdm1. Enteroendocrine cells are diploid and express the transcription factor Prospero (Pros). UAS-linked genes can be targeted to enterocytes by the MyolAGal4 (NP1–Gal4) driver8 or to ISCs and enteroblasts by the escargot (esg)–Gal4 driver2. To target expression of UAS-linked genes in RNSCs, we also used esg–Gal4 (ref. 4); for the quiescent HISCs, the wingless (wg)–Gal4 driver was used5,6. To investigate the response of the different cells to cell-death effectors, we first overexpressed reaper (rpr, an inhibitor of Death-associated inhibitor of apoptosis 1; Diap-1) in them, using the cell-type-specific Gal4 drivers combined with the temperature-sensitive Gal4 repressor tub–Gal80ts (ref. 29).
We used the inducible NP1–Gal4; tub–Gal80ts–UAS–rpr; UAS–GFP to express rpr in enterocytes (NP1ts > rpr), esg–Gal4; tubGal80ts–UAS–rpr; UAS–GFP (esgts > rpr) in ISCs, enteroblasts and RNSCs, and wg–Gal4; tubGal80ts–UAS–rpr; UAS–GFP (wgts > rpr) in HISCs. The NP1ts > rpr flies were raised to adulthood at 18 °C and shifted to 29 °C for 24 h to induce rpr expression.
Four male UAS–RNAi transgene flies were crossed with 8 female virgins of NP1ts, esgts, and wgts at 18 °C. Three- to five-day-old adult flies with the appropriate genotype were transferred to new vials at 29 °C for the indicated times before dissection. For p53, we did not find a significant change in esg+ progenitors and enteroendocrine cell numbers after the flies (esgts > p53) were cultured for 7 days at 29 °C, although a previous study found that a 15-day induction ablated nearly all esg+ cells and reduced enteroendocrine cell numbers8.
Histology and image capture
Fly intestines were dissected in PBS and fixed in PBS containing 4% formaldehyde for 30 min. After three 5-min rinses with 1× PBT (PBS + 0.1% Triton X-100), the samples were blocked in 1× PBT containing 5% normal goat serum overnight at 4 °C and incubated first with primary antibody overnight at 4 °C or at room temperature for 2 h, and then with a fluorescence-conjugated secondary antibody for 2 h at room temperature. Samples were mounted in Vectashield mounting medium with DAPI (Vector Laboratories). The following antibodies were used: rabbit polyclonal anti-β-Gal (1:1,000; Cappel); mouse anti-Dl (Delta, 1:20; DSHB); mouse monoclonal anti-Prospero (Pros, 1:50; DSHB); rabbit polyclonal anti-Pdm1 (1:1,000, a gift from X. Yang); mouse monoclonal anti-Arm N27A1 (1:20; DSHB); Rabbit monoclonal anti-Phospho-SAPK/JNK (1:200; Cell Signaling); rabbit-polyclonal anti-GFP (1:500, Invitrogen); mouse monoclonal anti-GFP (1:100; Invitrogen), and chicken polyclonal anti-GFP (1:3,000; Abcam). Secondary antibodies were goat anti-mouse, anti-chicken, and goat anti-rabbit IgG conjugated to Alexa488 or Alexa568 (1:400; Invitrogen). DAPI (Sigma) was used to stain DNA. CellMask Deep Red plasma membrane dye (Life Technologies, C10046) was used to visualize the plasma membrane. Midguts were labelled with CellMask Deep Red Plasma Membrane Stains (1:2,000) for 7 min30.
ROS detection by DHE
DHE staining was performed as described previously31. In brief, guts were dissected in 1× PBS, incubated in 30 μM DHE (Invitrogen) in PBS for 5 min at room temperature in the dark, washed twice, mounted and immediately imaged by confocal microscopy.
Lysotracker staining
Guts were dissected in 1× PBS and then stained without fixation in 0.5 μM Lysotracker Red DND-99 (Invitrogen) for 3 min at room temperature. They were then washed three times in 1× PBS, fixed for 20 min in 4% formaldehyde, washed three times in 1× PBT, rinsed twice with 1× PBS, mounted in Vectashield with DAPI and analysed on a confocal microscope.
Apoptosis detection
Apoptosis was detected by TUNEL with the ApopTag Red In situ Apoptosis Detection Kit (Chemicon International) according to the manufacturer’s instructions.
Necrosis evaluation by propidium iodide
Guts were dissected in 1× PBS and then stained in 1.5 μM PI (Invitrogen) for 15 min at room temperature. The guts were then fixed for 20 min in 4% formaldehyde, washed three times in 1× PBT, rinsed twice with 1× PBS, mounted in Vectashield with DAPI and analysed on a confocal microscope.
Oil Red O staining
Oil Red O staining was performed as described previously32. In brief, Drosophila midguts were dissected in 1× PBS and fixed in 4% formaldehyde for 30 min. Midguts were washed three times in 1× PBS, double-distilled water and a 60% isopropanol solution. From the stock solution of Oil Red O (Sigma-Aldrich; 0.1% solution in isopropanol), a working solution was prepared by mixing 6 ml of 0.1% Oil Red O in isopropanol and 4 ml of double-distilled water. Midguts were incubated for 20 min in this solution and then washed in 60% isopropanol and water. The midguts were mounted in Vectashield mounting medium with DAPI (Vector Laboratories) and were imaged by confocal microscopy. Images were captured with the Zeiss LSM 510 confocal system and processed with LSM Image Browser and Adobe Photoshop.
Quantification and statistical analysis
To determine the percentage of GFP+ cells, the GFP+ cells and total cells were counted in a 5,000-μm2 area of a single confocal plane. In esgts samples, cells were counted in the posterior midgut and Malpighian tubules; in wgts samples, they were counted in the hindgut–midgut junction; and in esgtswgts samples, they were counted in the hindgut–midgut junction, the posterior midgut and the Malpighian tubules. The number of Pros+ nuclei was counted in a 0.08-mm2 surface area of a microscopic image from a similar region of each posterior midgut33. Cells per tumour were determined by counting the total number of nuclei within GFP+ tumours. All of the images were taken with the LSM5 Image Browser using the same confocal settings (Zeiss). Statistical analyses were performed using GraphPad Prism. Sample sizes (n) reported reflect the number of individual midguts. All experiments were performed in triplicate. P values were obtained between two groups using the Student’s t-test. For all statistical analysis, differences were considered to be statistically significant at values of P < 0.05.
Flow cytometry
Cell surface markers were analysed by flow cytometry. Cultured or treated cells were dissociated by 0.05% trypsin-EDTA and centrifuged. Single cells were resuspended with PBS containing 2% FBS and fluorescent-conjugated antibodies FITC-CD44 (clone G44-26, BD Biosciences) and PE-CD24 (clone ML5, Biolegend), and incubated on ice for 30 min. After washing three times with PBS containing 2% FBS, cells were resuspended with PBS containing 2% FBS and analysed by BD FACS Caliber (BD Biosciences).
Inhibitors
Arf1 inhibitors are BFA (brefeldin A)34 from Sigma, GCA (golgicide A)35 from Santa Cruz, secin H336 from Cayman chemical, LM1137 from A. Chavanieu, LG838 from L. Frigerio. 2-deoxy-d-Glucose (2-DG)39; JNK inhibitor SP 60012540; FAO inhibitors:triascin C41, etomoxir42, and mildronate43 from Cayman chemical, Enoximone44 from Tocris. Rac1 inhibitor from Santa Cruz. N-Acetyl-L-cysteine (NAC) 45 from Sigma. For control experiments, a DMSO control (100 μl in 10 ml food) was used. All inhibitors were mixed in the fly food with following concentrations; Arf1 inhibitors: BFA (50 ng ml−1 and 200 ng ml−1), GCA (5 μM), LM11 (50 μM), LG8 (100 μM), and secin H3 (50 μM); JNK inhibitor: Sp600125 (50 μM); Rac1 inhibitor (100 μM); ROS inhibitor: NAC (10 mM); FAO inhibitors: triacsin C (5 μM), mildronate (100 μM), etomoxir (100 μM), enoximone (100 μM); and glycolysis inhibitor 2-DG (50 mM). We mixed each inhibitor in fly food and tested different concentrations of these inhibitors. We used the concentration in which the inhibitors could kill tumour cells. At the beginning of experiments, to find out whether the flies would eat the inhibitors, we added green food dye to fly food. Flies were fasted for 1 h and then 8–10 flies were transferred to a vial containing coloured fly food mixed with inhibitors. We found that within 1 h of feeding the green dye could be seen through the abdomen of each fly, which suggest that the fly food mixed with inhibitors was edible to the flies. However, we excluded the food dye from the food used in the main experiments. For the main experiments, we fed flies with food containing inhibitors for 4 days. We repeated each inhibitor treatment three times.
Cell lines and culture
The human prostate cancer cell line DU145, colon cancer cell line HT29, and breast cancer cell lines MCF7, MDA-MB-231 (provided by the DCTD Tumour Repository) were cultured in RPMI1640 supplemented with 10% fetal bovine serum and 100 units per ml penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2.
Cell survival assay
Cells were seeded at 2 × 105 cells per well in 6-well plates. Treatments with the indicated chemicals were started the next day, and cells were incubated for 2 more days. The surviving cells were stained with Crystal Violet (EMD Millipore). For quantification, the stained cells were solubilized in 1% SDS, and the absorbance at 595 nm was determined with a microplate reader.
Sphere formation assay
Single cells were cultured in a Corning Costar Ultra-Low attachment 24-well plate (Sigma-Aldrich) in sphere culture medium, consisting of DMEM/F12 (1:1), B27 (Invitrogen), and 20 ng ml−1 EGF (Invitrogen), with the indicated chemicals. The number of spheres was counted after 10 days of culture.
RNA isolation and real-time PCR
An RNeasy Mini Kit (Qiagen) was used to extract the total RNA from human cancer cells. The cDNA was synthesized from 1 μg RNA from each sample using a reverse transcription kit (Promega). Real-time PCR was performed in a 15-μl reaction system using SYBR Advantage qPCR Premix (Clontech). All of the reactions were performed in triplicate in a RealPlex 2 system (Eppendorf). The relative gene expression was quantified as described previously46. The sequence of each primer was as follows: ACTB, 5′-GATCATTGCTCCTCCTGAGC-3′ and 5′-ACTCCTGCTTGCTGATCCAC-3′;CDH1, 5′-ACCAGAATAAAGACCAAGTGACCA-3′ and 5′-AGCAAGAGCAGCAGAATCAGAAT-3′; CD44, 5′-GAGCATCGGATTTGAGA-3′ and 5′-CATACTGGGAGGTGTTGG-3′.
References
Kleffel, S. & Schatton, T. Tumor dormancy and cancer stem cells: two sides of the same coin? Adv. Exp. Med. Biol. 734, 145–179 (2013)
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006)
Ohlstein, B. & Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474 (2006)
Singh, S. R., Liu, W. & Hou, S. X. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell 1, 191–203 (2007)
Fox, D. T. & Spradling, A. C. The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 5, 290–297 (2009)
Takashima, S., Mkrtchyan, M., Younossi-Hartenstein, A., Merriam, J. R. & Hartenstein, V. The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 454, 651–655 (2008)
Essers, M. A. G. & Trumpp, A. Targeting leukemic stem cells by breaking their dormancy. Mol. Oncol. 4, 443–450 (2010)
Jiang, H. et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 (2009)
Ma, M. et al. Wildtype adult stem cells, unlike tumor cells, are resistant to cellular damages in Drosophila. Dev. Biol. 411, 207–216 (2016)
Zeng, X., Singh, S. R., Hou, D. & Hou, S. X. Tumor suppressors Sav/Scrib and oncogene Ras regulate stem-cell transformation in adult Drosophila malpighian tubules. J. Cell. Physiol. 224, 766–774 (2010)
Ohlstein, B. & Spradling, A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992 (2007)
Goulas, S., Conder, R. & Knoblich, J. A. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529–540 (2012)
Wu, M., Pastor-Pareja, J. C. & Xu, T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature 463, 545–548 (2010)
Lee, M. C. S., Miller, E. A., Goldberg, J., Orci, L. & Schekman, R. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20, 87–123 (2004)
Beller, M. et al. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6, e292 (2008)
Zeng, X. et al. Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Reports 10, 1226–1238 (2015)
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999)
Zhang, Y., Chen, D. & Wang, Z. Analyses of mental dysfunction-related ACSl4 in Drosophila reveal its requirement for Dpp/BMP production and visual wiring in the brain. Hum. Mol. Genet. 18, 3894–3905 (2009)
Min, K. T. & Benzer, S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science 284, 1985–1988 (1999)
Grönke, S. et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323–330 (2005)
Palanker, L., Tennessen, J. M., Lam, G. & Thummel, C. S. Drosophila HNF4 regulates lipid mobilization and β-oxidation. Cell Metab. 9, 228–239 (2009)
Timmons, A. K., Meehan, T. L., Gartmond, T. D. & McCall, K. Use of necrotic markers in the Drosophila ovary. Methods Mol. Biol. 1004, 215–228 (2013)
Etchegaray, J. I. et al. Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development 139, 4029–4039 (2012)
Ohsawa, S. et al. Elimination of oncogenic neighbors by JNK-mediated engulfment in Drosophila. Dev. Cell 20, 315–328 (2011)
Ohashi, Y. et al. AMF-26, a novel inhibitor of the Golgi system, targeting ADP-ribosylation factor 1 (Arf1) with potential for cancer therapy. J. Biol. Chem. 287, 3885–3897 (2012)
Trumpp, A. & Wiestler, O. D. Mechanisms of disease: cancer stem cells—targeting the evil twin. Nat. Clin. Pract. Oncol. 5, 337–347 (2008)
Han, C. Z. & Ravichandran, K. S. Metabolic connections during apoptotic cell engulfment. Cell 147, 1442–1445 (2011)
McPhee, C. K., Logan, M. A., Freeman, M. R. & Baehrecke, E. H. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature 465, 1093–1096 (2010)
McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. & Davis, R. L. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 (2003)
Hsieh, H. H., Chang, W. T., Yu, L. & Rao, Y. Control of axon-axon attraction by Semaphorin reverse signaling. Proc. Natl. Acad. Sci. USA 111, 11383–11388 (2014)
Hochmuth, C. E., Biteau, B., Bohmann, D. & Jasper, H. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8, 188–199 (2011)
Reiff, T. et al. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. eLife 4, e06930 (2015)
Amcheslavsky, A. et al. Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Reports 9, 32–39 (2014)
Sausville, E. A. et al. Antiproliferative effect in vitro and antitumor activity in vivo of brefeldin A. Cancer J. Sci. Am. 2, 52–58 (1996)
Sáenz, J. B. et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5, 157–165 (2009)
Hafner, M. et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature 444, 941–944 (2006)
Viaud, J. et al. Structure-based discovery of an inhibitor of Arf activation by Sec7 domains through targeting of protein-protein complexes. Proc. Natl Acad. Sci. USA 104, 10370–10375 (2007)
Sorieul, M. et al. An Exo2 derivative affects ER and Golgi morphology and vacuolar sorting in a tissue-specific manner in arabidopsis. Traffic 12, 1552–1562 (2011)
Ciavardelli, D. et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 5, e1336 (2014)
Bennett, B. L. et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl Acad. Sci. USA 98, 13681–13686 (2001)
Mashima, T. et al. p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target. J. Natl. Cancer Inst. 97, 765–777 (2005)
Samudio, I. et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Invest. 120, 142–156 (2010)
Dambrova, M., Liepinsh, E. & Kalvinsh, I. Mildronate: cardioprotective action through carnitine-lowering effect. Trends Cardiovasc. Med. 12, 275–279 (2002)
Abdel-Aleem, S., Youssef, J., Frangakis, C. & Badr, M. Selective inhibition of hepatic peroxisomal fatty acid beta-oxidation by enoximone. Life Sci. 51, 53–57 (1992)
Delneste, Y., Jeannin, P., Potier, L., Romero, P. & Bonnefoy, J. Y. N-acetyl-L-cysteine exhibits antitumoral activity by increasing tumor necrosis factor alpha-dependent T-cell cytotoxicity. Blood 90, 1124–1132 (1997)
Feng, X. et al. Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J. Biotechnol. 150, 288–293 (2010)
Singh, S. R., Zeng, X., Zheng, Z. & Hou, S. X. The adult Drosophila gastric and stomach organs are maintained by a multipotent stem cell pool at the foregut/midgut junction in the cardia (proventriculus). Cell Cycle 10, 1109–1120 (2011)
Strand, M. & Micchelli, C. A. Quiescent gastric stem cells maintain the adult Drosophila stomach. Proc. Natl Acad. Sci. USA 108, 17696–17701 (2011)
O’Brien, L. E., Soliman, S. S., Li, X. & Bilder, D. Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603–614 (2011)
Kirilly, D., Spana, E. P., Perrimon, N., Padgett, R. W. & Xie, T. BMP signaling is required for controlling somatic stem cell self-renewal in the Drosophila ovary. Dev. Cell 9, 651–662 (2005)
Adachi-Yamada, T., Fujimura-Kamada, K., Nishida, Y. & Matsumoto, K. Distortion of proximodistal information causes JNK-dependent apoptosis in Drosophila wing. Nature 400, 166–169 (1999)
Zhou, Z., Hartwieg, E. & Horvitz, H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43–56 (2001)
Ziegenfuss, J. S., Doherty, J. & Freeman, M. R. Distinct molecular pathways mediate glial activation and engulfment of axonal debris after axotomy. Nat. Neurosci. 15, 979–987 (2012)
Fadok, V. A. et al. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405, 85–90 (2000)
Wu, Y., Tibrewal, N. & Birge, R. B. Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 16, 189–197 (2006)
Dontu, G. et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17, 1253–1270 (2003)
Acknowledgements
We thank S. Hayashi, J.-P. Vincent, M. Fortini, C. Thummel, E. Baehrecke, R. P. Kuhnlein, M. Freeman, F. Schweisguth, M. Mlodzik, T. Lecuit, DGRC, VDRC, and the Bloomington Stock Centers for fly stocks; A. Chavanieu and L. Frigerio for Arf1 inhibitors; X. Yang and the Developmental Studies Hybridoma Bank for antibodies; and S. Lockett for help with the confocal microscope. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Author information
Authors and Affiliations
Contributions
S.X.H., X.Z. and S.R.S. conceived and designed the experiments. S.R.S., X.Z. and S.X.H. performed the Drosophila experiments. J.Z. performed the experiments using human cancer cell lines. Y.L., G.H. and H.L. assisted with experiments. S.X.H., S.R.S., X.Z. and J.Z. analysed the data. S.X.H., S.R.S. and J.Z. wrote the manuscript. S.X.H. and S.R.S. revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks Y. Apidianakis, M. Montminy, H. Steller and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Stem cells are resistant to apoptosis.
a, Stem cells in the adult Drosophila digestive system. In this system, three organs, the posterior midgut, the hindgut and the Malpighian tubules, meet and join at the junction of the posterior midgut and hindgut. Stem cells in these organs exhibit different degrees of quiescence. The intestinal stem cells (ISCs), located in the posterior midgut, divide once every 24 h2,3; the renal and nephric stem cells (RNSCs), located in the Malpighian tubules, divide about once a week4; and the quiescent hindgut intestinal stem cells (HISCs), found at the midgut–hindgut junction, divide only during stress-induced tissue repair5,6. GaSCs are gastric stem cells at the foregut–midgut junction47. GSSCs are gastric stem cells in the middle of the midgut48. The colours just make the cell types or organs more visible and do not exactly reflect different regions in the digestive system. b–n Stem cells are resistant to apoptosis. b, NP1ts > rpr, 18 °C, 24 h (n = 37). c, NP1ts > rpr, 29 °C, 24 h (n = 29). d, e, esgts > lacZRNAi, 29 °C, 7 d (n = 32). f, g, esgts >rpr, 29 °C, 7 d (n = 35). h, wgts > lacZRNAi, 29 °C, 7 d (n = 27). i, wgts > rpr, 29 °C, 7 d (n = 24). j, NP1ts > p53, 29 °C, 5 d (n = 31). k, esgts > p53, 29 °C, 7 d (n = 38). l, Quantification of Apoptag+ cells in the indicated panels. m, Quantification of GFP+ cells in the indicated panels. n, Quantification of Pros+ cells in the indicated panels. Data are represented as mean ± s.d.. Statistical significance determined by Student’s t-test, ***P < 0.0001. NS, not significant (P > 0.05). As reported previously8, 24-h induction of rpr in enterocytes resulted in widespread apoptosis (compare c with b and see the quantitative comparison in l). The induction of rpr by esg-Gal4 (f, g) or wg-Gal4 (i) had little effect on the progenitor or stem cells (that is, enteroblasts, ISCs, RNSCs and HISCs) at one week, compared to wild-type controls (compare f, g with d, e; i with h, and see the quantitative comparison in m). We also found that the overexpression of Drosophila p53 could effectively ablate the enterocytes in five days (compare j with b and see the quantitative comparison in l) but had little effect on stem cells even after one week, compared to controls (compare k with e and see the quantitative comparison in m). Because NP1–Gal4 and esg–Gal4 are not expressed in enteroendocrine cells, as expected, we did not find significant changes in enteroendocrine cells in these experiments (n). o–u, Activation of proliferation accelerates apoptotic cell death of hyperplastic stem cells but fails to completely eliminate neoplastic stem cells. o, Quantification of Apoptag+ cells in the indicated panels. Data are represented as mean ± s.d. Statistical significance was determined by Student’s t-test, ***P < 0.0001. p, esgts > upd, 29 °C, 4 d (n = 28). q, esgts > upd + rpr, 29 °C, 4 d (n = 33). r, s, esgts > NDN, 29 °C, 7 d (n = 25). t, u, esgts > NDN + rpr, 29 °C, 7 d (n = 32). White arrows point to the hindgut–midgut junction in h, i, p, q; yellow arrows point to Pros+ enteroendocrine cells in r and t; green arrows point to Dl+ ISCs in r and t. White dotted lines outline GFP+ stem cell clusters in r and t. Yellow dotted lines outline enteroendocrine cell clusters in r and t. Expression of rpr or Arf79FRNAi in ISCs did not kill differentiated cells. The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies and analysed by confocal microscopy. Scale bars in b–k and p–u, 10 μm.
Extended Data Figure 2 The COPI–Arf1 complex regulates stem but not differentiated cell survival.
a, The COPI–lipolysis–β-oxidation pathway. The COPI–Arf1 complex controls lipid homeostasis by regulating adipocyte differentiation-related protein (ADRP), tail-interacting protein of 47kDa (Tip47) and adipocyte triglyceride lipase (ATGL)15. Triglycerides (TG), diglyceride (DG), fatty acid (FA), Acyl-CoA synthetase (ACS). b–j, The COPI–Arf1 complex regulates stem cell survival. The genotypes of the flies in each panel were: b, esgts wgts > lacZRNAi, 29 °C, 7 d (n = 38). c, esgts wgts > β-CopRNAi, 29 °C, 7 d (n = 23). d, esgts wgts > δ-CopRNAi, 29 °C, 7 d (n = 32). e, esgts wgts > γ-CopRNAi, 29 °C, 7 d (n = 27). f, esgts wgts > ζ-CopRNAi, 29 °C, 7 d (n = 31). g, esgts wgts > β’-CopRNAi, 29 °C, 7 d (n = 29). h, esgts wgts > garzRNAi, 29 °C, 7 d (n = 27). i, esgts wgts > AcslRNAi, 29 °C, 7 d (n = 32). j, esgts wgts >bgmRNAi, 29 °C, 7 d (n = 25). k, Quantification of GFP+ cells in the indicated panels. Data show mean ± s.e.m. Statistical significance was determined by Student’s t-test, ***P < 0.0001. NS, not significant (P > 0.05). l–q Knockdown of the COPI–Arf79F complex did not kill differentiated cells. The genotypes of the flies in each panel were: l, n, NP1ts > lacZRNAi, 29 °C, 7 d (n = 25). m, o, NP1ts > Arf79FRNAi, 29 °C, 7 d (n = 32). p, tshts >lacZRNAi, 29 °C, 7 d (n = 22). q, tshts > Arf79FRNAi, 29 °C, 7 d (n = 25). r, esgts wgts > RasV12 + ζ-CopRNAi, 29 °C, 7 d (n = 27). s, esgts wgts > NDN + δ-CopRNAi, 29 °C, 7 d (n = 30). t, u, esgts > NDN + Arf79FRNAi, 29 °C, 7 d (n = 40). v, esgts wgts > RasV12 + AcslRNAi, 29 °C, 7 d. (w) esgts wgts > β-CopRNAi + Hnf4, 29 °C, 7 d. x, esgts wgts > ζ-CopRNAi + scu, 29 °C, 7 d. White arrows in b–j and r, s, w, x, point to the hindgut–midgut junction. Yellow arrows point to Pros+ enteroendocrine cells in t; green arrows point to Dl+ ISCs in t, and a white arrow points to a remaining GFP+ stem cell in u. Scale bars in b–j and l–x, 10 μm.
Extended Data Figure 3 The COPI–Arf79F complex regulates stem cell survival through lipolysis and knockdown of these genes blocks lipolysis, but promotes lipid storage.
a–h, The COPI complex autonomously regulates stem cell survival. The three- or four-day-old adult female flies were heat-shocked twice with an interval of 8–12 h, at 37 °C, for 60 min to induce MARCM clones17. In wild-type clones, small GFP+ cell clusters were detected 2 d ACI (a, h; n = 33), which grew into large clusters that contained both ISCs and their differentiated progenies by 7 d ACI (b, h; n = 37). In the δ-COP mutant clones, only a few GFP+ cells were identified 2 d ACI (c, h; n = 34), and none were seen at 7 d ACI (f, h; n = 31). Similarly, only a few GFP+ cells were identified at 2 d ACI in γ-COP (e, h; n = 27) mutant clones, and none were seen at 7 d ACI (f; h; n = 34). Expressing UAS–γ-COP–GFP in γ-COP10-mutant MARCM clones (g and h; n = 31) completely rescued the stem cell death phenotype. These results suggest that the COPI complex cell-autonomously regulates stem cell survival. Dotted lines in a and b outline GFP+ clones. White arrows in c and e point to individual GFP+ cells. h, Quantification of GFP+ cells in the indicated panels. Data show the mean ± s.e.m. Statistical significance was determined by Student’s t-test, ***P < 0.0001. The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies and analysed by confocal microscopy. i–p, The lipolysis pathway is active in stem cells. To further investigate the function of lipolysis in stem cells, we investigated the expression of a lipolysis reporter (GAL4–dHFN4; UAS–nlacZ)21. In our system, this reporter showed strong β-galactosidase expression in mira-GFP-positive ISCs and RNSCs (i–k, n = 15), but not in enterocytes, enteroendocrine cells, and the quiescent HISCs of mature adult flies (i, white arrows, 3–5 days old) or in the quiescent ISCs of freshly emerged young adult flies (less than 3 days old; l and m, n = 17)49 at 29 °C culture conditions. Expressing δ-COPRNAi (esgts > δ-COPRNAi + GAL4–dHFN4; UAS–nlacZ) almost completely eliminated the reporter expression (n, n = 24), suggesting that the reporter is specifically regulated by the COPI complex. We also expressed a constitutively active form of JAK (hopTum-l) with GAL4–dHFN4; UAS–nlacZ and found that the reporter was expressed in hopTum-l-activated HISCs (o, white arrows, n = 20). The GAL4 in the reporter system is under the control of an hsp70 promoter; we heat-shocked the flies for 30 min at 37 °C 12 h before dissection and found that the reporter was strongly expressed in ISCs, RNSCs and HISCs (particularly strong in HISCs), but not in enteroendocrine cells and enterocytes (p, white arrows, n = 17). Arrows in i, n, o and p point to HISCs at the hindgut–midgut junction. q–v, Arf79F knockdown promotes lipid storage in stem cells. The genotypes of the flies in each panel were: q–s, esgts > lacZRNAi, 29 °C, 4 d (n = 30). t–v, esgts > Arf79FRNAi, 29 °C, 4 d (n = 37). The posterior midguts of flies with the indicated genotypes were dissected, stained with Oil Red O (red), anti-GFP (green) and DAPI (blue), and analysed by confocal microscopy. Dotted lines outline stem cells and white arrows point to lipid droplets in stem cells. Scale bars in a–g and i–v, 10 μm.
Extended Data Figure 4 The lipolysis–β-oxidation pathway regulates survival of transformed stem cells.
a–l, Arf1 inhibitors kill RasV12-transformed RNSCs through the ROS–Rac–JNK pathway. The GFP-labelled RNSC tumour clusters were induced by expressing RasV12 in RNSC clones, using the positively marked mosaic lineage (PMML) labelling technique10,50 in adult Drosophila. The flies with RasV12-PMML clones were cultured for 4 d at room temperature on normal food to let the tumour grow and then switched to food with indicated drugs for another 4 d. Flies with RasV12-tumours were given normal food with DMSO (a), 50 ng ml−1 BFA (b), 5 μM GCA (c), 50 μM LM11 (d), 100 μM LG8 (e), 50 μM secin H3 (f), 50 μM secin H3 + 50 μM JNK inhibitor Sp600125 (g), 50 μM secin H3 + 100 μM Rac1 inhibitor (h) or 50 μM secin H3 + 10 mM NAC (i). j,k, esgts flies were fed with normal food with either DMSO (j, n = 20) or 50 μM secin H3 (k, n = 22). n = number of tissues observed. l, Quantification analysis of tumour sizes in Malpighian tubules of indicated panels. We classified all tumours into four categories based on the total number of GFP+ cells in each tumour clone (<10 cells, 10–20 cells, 20–50 cells and 50–100 cells). Total number of tumours investigated for each treatment: DMSO (466 tumours, n = 27 Malpighian tubules), BFA (63 tumours, n = 30 Malpighian tubules), GCA (73 tumours, n = 32 Malpighian tubules), LM11 (94 tumours, n = 35 Malpighian tubules), LG8 (86 tumours, n = 27 Malpighian tubules), secin H3 (64 tumours, n = 25 Malpighian tubules), secin H3 + JNK inhibitor (220 tumours, n = 30 Malpighian tubules), Secin H3 + Rac1 inhibitor (211 tumours, n = 27 Malpighian tubules), and Secin H3 + NAC (297 tumours, n = 35 Malpighian tubules). Arrows point to GFP+ RNSC tumour clusters in a–i. m–r, The lipolysis pathway regulates survival of transformed stem cells. The genotypes of the flies in each panel were: m, m′, esgts > NDN, 29 °C, 4 d (m, n = 30; m′, n = 35). n, n′, esgts > NDN + Hnf4, 29 °C, 4 d (n, n = 25; n′, n = 27). o, o ′, esgts > RasV12, 29 °C, 4 d (o, n = 25; o′, n = 32). p, p′, esgts > RasV12 + Hnf4, 29 °C, 4 d (p, n = 25; p′, n = 32). q, q′, esgts > RasV12 + scu, 29 °C, 4 d (q, n = 23; q′, n = 30). The flies were fed with normal food with either DMSO (m–q) or BFA (m′ and n′, 200 ng ml−1; o′–q′, 50 ng ml−1) for 4 d. Expressing Hnf4 or scu partially blocked the effect of BFA on transformed stem cells. r, Quantification of esg > GFP+ tumour cells in 5 × 103 μm2 per treatment in indicated panels. Data show the mean ± s.e.m. Statistical significance was determined by Student’s t-test, ***P < 0.0001. NS, not significant (P > 0.05). Arrows point to GFP+ RNSC tumour clusters in o–q′. s–y, FAO inhibitors, but not 2-DG, kill RasV12-transformed RNSCs. The GFP-labelled RNSC tumour clusters were induced by expressing RasV12 in RNSC clones using the PMML technique in adult Drosophila. The flies with RasV12-PMML clones were cultured for 4 d at room temperature on normal food to let the tumour grow and then switched to food with indicated drugs for another 4 d. Flies with RasV12-tumours were given normal food with DMSO (s), 5 μM triacsin C (t), 100 μM mildronate (u, n = 27), 100 μM etomoxir (v), 100 μM enoximone (w, n = 37) or 50 mM 2-deoxyglucose (2-DG) (x, n = 32). y, Quantification analysis of tumour sizes in Malpighian tubules of indicated panels. Total number of tumours investigated for each treatment: DMSO (474 tumours, n = 30 Malpighian tubules), triacsin C (47 tumours, n = 32 Malpighian tubules), mildronate (69 tumours, n = 27 Malpighian tubules), etomoxir (73 tumours, n = 35 Malpighian tubules), enoximone (86 tumours, n = 27 Malpighian tubules) and 2-DG (264 tumours, n = 32 Malpighian tubules). Arrows point to GFP+ RNSC tumour clusters. The gut of flies with the indicated genotypes was dissected after cultured, stained with the indicated antibodies and analysed by confocal microscopy. Scale bars in a–k, m–q’ and s–x, 10 μm.
Extended Data Figure 5 Knockdown of components of the COPI–Arf79F–Acsl pathway kill normal and transformed stem cells through necrosis.
a–i, The genotypes of the flies in each panel were: a–c and f–i, esgts > Arf79FRNAi, 29 °C, 7 d (n = 27). d, e, esgts > lacZRNAi, 29 °C, 7 d (n = 20). In d–i, a dye (CellMask) marks plasma membranes. In a-c and g-i, a dying ISC is engulfed by a neighbouring enterocytes. j, esgts wgts > lacZRNAi, 29 °C, 4 d (n = 30). k, esgts wgts > ζ-copRNAi, 29 °C, 4 d (n = 36). l, esgts wgts> β-CopRNAi, 29 °C, 4 d (n = 34). m, esgts wgts > garzRNAi, 29 °C, 4 d (n = 32). n, o, esgts wgts > AcslRNAi, 29 °C, 4 d (n = 32). p, Quantification of propidium-iodide-positive cells in the indicated panels. Data show the mean ± s.d. Statistical significance was determined by Student’s t-test, ***P < 0.0001 (compared to control). Yellow arrows point to hindgut–midgut junctions in j and n, white arrows point to GFP- and propidium-iodide-positive stem cells in k and o. The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies or reagents and analysed by confocal microscopy. Scale bars in a–o: 10 μm.
Extended Data Figure 6 Arf79F knockdown kills transformed and normal stem cells through necrosis.
(a–d) The genotypes of the flies in each panel were: a, esgts > NDN, 29 °C, 4 d (n = 25). b, c, esgts > NDN + Arf79FRNAi, 29°C, 4 d (n = 25). d, Quantification of PI+ cells in the indicated panels. Data show the mean ± s.d. Statistical significance was determined by Student’s t-test, ***P < 0.0001. NS, not significant (P > 0.05). White arrows point to GFP- and propidium-iodide-positive stem cells in c. e–l, The genotypes of the flies in each panel were: e, f, i, j, esgts > lacZRNAi, 29 °C, 7 d (e, f, n = 27; i, j, n = 30). g, h, k, l, esgts > Arf79FRNAi, 29 °C, 7 d (g, h, n = 45; k, l, n = 30). White arrows point to GFP+ stem cells. No DHE or LysoTracker signals were detected in the wild-type midgut, but these signals were intense in the esgts > Arf79FRNAi flies, indicating that the dying ISCs had accumulated ROS and were intracellularly acidified. The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies or dyes and analysed by confocal microscopy. Scale bars in a–c and e–l: 10 μm.
Extended Data Figure 7 Overexpressing Cat rescues the ISC death induced by Arf79FRNAi but not CycTRNAi expression and dying ISCs activate JNK signalling and autophagy in enterocytes.
a–h, The genotypes of the flies in each panel were: a, esgts > Cat, 29 °C, 7 d (n = 25). b, esgts > Arf79FRNAi + Cat, 29 °C, 7 d (n = 32). c, esgts > Sod, 29 °C, 7 d (n = 24). d, esgts > Arf79FRNAi + Sod, 29 °C, 7 d (n = 30). e, esgts > Sod2, 29 °C, 7 d (n = 22). f, esgts > Arf79FRNAi + Sod2, 29 °C, 7 d (n = 32). g, esgts > CycTRNAi, 29 °C, 7 d (n = 35). h, esgts > CycTRNAi + Cat, 29 °C, 7 d (n = 37). Overexpressing Cat, but not sod or sod2, in stem cells (esgts > Arf79FRNAi + Cat) rescued the stem-cell death induced by Arf79F knockdown but not that induced by CycT knockdown (esgts>CycTRNAi+Cat). i–n, The genotypes of the flies in each panel were: i, puc–lacZ, 29 °C, 7 d (n = 17). j, esgts > Arf79FRNAi + puc–lacZ, 29 °C, 7 d (n = 20). Yellow arrows point to GFP+ cells. k, esg > GFP, 29 °C, 4 d (n = 12). l, FRT82B–γ-COP10 MARCM clones, 4 d (n = 17). Yellow arrows point to GFP+ clones. m, esgts > lacZRNAi + pmCherry–Atg8a, 29 °C, 7 d (n = 22). n, esgts > Arf79FRNAi + pmCherry–Atg8a, 29 °C, 7 d (n = 25). Arf79F knockdown in ISCs induced Puc–lacZ (compare i with j) and Cherry–Atg8a (compare m with n) expression in enterocytes, p-JNK was induced in enterocytes in γ-COP mutant MARCM clones, compare k with l). The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies and analysed by confocal microscopy. Scale bars in a–n: 10 μm.
Extended Data Figure 8 Knockdown of components of the JNK pathway or engulfment genes in ISCs did not block the ISC death induced by Arf79FRNAi or δ-CopRNAi expression.
a–i, The genotypes of the flies in each panel were: a, esgts > Arf79FRNAi, 29 °C, 7 d (n = 27). b, esgts > δ-CopRNAi + Rac1DN, 29 °C, 7 d (n = 32). c, esgts > δ-CopRNAi + mbcRNAi, 29 °C, 7 d (n = 25). d, esgts > Arf79FRNAi + bskDN, 29 °C, 7 d (n = 30). e, esgts > Arf79FRNAi + drprRNAi, 29 °C, 7 d (n = 28). f, esgts > δ-CopRNAi + Atg5RNAi, 29 °C, 7 d (n = 32). g, esgts > Arf79FRNAi + p35, 29 °C, 7 d (n = 22). h, esgts > δ-CopRNAi + PSRRNAi, 29 °C, 7 d (n = 30). i, esgts > Arf79FRNAi + mysRNAi, 29 °C, 7 d (n = 28). bskDN is a dominant-negative form of Drosophila JNK (ref. 51), draper (drpr) encodes a homologue of the C. elegans transmembrane phagocytic receptor (ref. 52), Rac1 encodes a small GTPase that is a homologue of the C. elegans engulfment gene ced-10 (ref. 53), myoblast city (mbc)/Crk/dCed-12 encodes a Rac1 guanine nucleotide exchange factor (GEF) (ref. 53), PSR encodes a phosphatidylserine receptor (ref. 54) and mys encodes the β-subunit of integrin, which is involved in mammalian cell engulfment (ref. 55). Light chain 3 (LC3) in autophagosomes is involved in the rapid degradation of the internalized cargo (reviewed in Han and Ravichandran in ref. 27). j–l, Activation of hep or Rac1 genes in enterocytes induced the ISC death. The genotypes of the flies in each panel were: j, mira–GFP, 29 °C, 7 d (n = 17). k, mira–GFP + NP1ts (–UAS–GFP) > hepCA (a constitutively activate form of hep), 29 °C, 7 d (n = 15). l, mira–GFP + NP1ts (–UAS–GFP) > Rac1V12 (a constitutively activate form of Rac1), 29 °C, 3 d (n = 12). m–n, Overexpression of drpr in enterocytes did not induce EC death. m, NP1ts > lacZRNAi, 29 °C, 5 d (n = 15). n, NP1ts > drpr, 29 °C, 5 d (n = 20). The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies and analysed by confocal microscopy. Scale bars in a–n: 10 μm.
Extended Data Figure 9 Knockdown of components of the JNK pathway or engulfment genes in enterocytes blocks the ISC death induced by Arf79FRNAi or δ-CopRNAi expression.
The genotypes of the flies in each panel were: a, NP1ts esgts > lacZRNAi, 29 °C, 7 d (n = 20). b, NP1ts esgts > Arf79FRNAi, 29 °C, 7 d (n = 32). c, NP1ts esgts > δ-CopRNAi, 29 °C, 7 d (n = 30). d, NP1ts esgts > Arf79FRNAi + bskDN, 29 °C, 7 d (n = 30). e, NP1ts esgts > δ-CopRNAi + Rac1DN, 29 °C, 7 d (n = 202). f, NP1ts esgts > δ-CopRNAi + mbcRNAi, 29 °C, 7 d (n = 18). g, NP1ts esgts > Arf79FRNAi + drprRNAi, 29 °C, 7 d (n = 32). h, NP1ts esgts > δ-CopRNAi + Atg5RNAi, 29 °C, 7 d (n = 17). i NP1ts esgts > δ-CopRNAi + Atg12RNAi, 29 °C, 7 d (n = 25). j, NP1ts esgts > δ-CopRNAi + PSRRNAi, 29 °C, 7 d (n = 22). k, NP1ts esgts > Arf79FRNAi + mysRNAi, 29 °C, 7 d (n = 35). l, NP1ts esgts > Arf79FRNAi + p35, 29 °C, 7 d (n = 27). m, NP1ts > JraAsp (a constitutively activate form of Jun), 29 °C, 7 d (n = 25). n, NP1ts > drpr, 29 °C, 7 d (n = 20). Expression of JraAsp and drpr in enterocytes eliminates Dl+ ISCs. The posterior midguts of flies with the indicated genotypes were dissected, stained with the indicated antibodies and analysed by confocal microscopy. White arrows point to Dl+ ISCs, yellow arrowheads point to Pros+ enteroendocrine cells and green arrows point to enterocytes. Scale bars in a–n: 10 μm.
Extended Data Figure 10 Arf1 and FAO inhibitors suppress CSCs in human cancer cell lines.
(a–f) Arf1 inhibitors suppress proliferation and sphere formation in DU145 cells. a, b, Crystal violet staining was used to detect cell survival after 2 days of treatment with BFA or GCA at the indicated concentrations in DU145 cells. The growth of DU145 cells was strongly inhibited by 30 ng ml−1 BFA (a, b) and 2.5 μM GCA (b). We also tested the two inhibitors in tumour sphere formation by cancer cells, a widely used in vitro technique for assessing CSC self-renewal capacity56. Spheres were cultured with or without BFA or GCA. The two inhibitors also inhibited tumour sphere formation (c, d). GCA was a weak inhibitor of growth (b), but a strong inhibitor of tumour sphere formation (d) in DU145 cells, indicating that these inhibitors may specifically target CSCs. The mRNA level of CD44 and E-Cadherin, two potent prostate cancer tumour-initiating cell markers, were reduced by BFA treatment (e, f). Data show the mean ± s.e.m. Statistical significance was determined by Student’s t-test, *P < 0.05; **P < 0.01 (compared to DMSO). g–l, Arf1 inhibitors suppress proliferation and sphere formation in HT29 and MCF7 cells. Crystal violet staining was used to detect cell survival after 2 days of treatment with BFA or GCA at the indicated concentrations in HT29 and MCF7 cells. The inhibitors reduced the cell survival rate (g–j). Spheres were cultured with or without BFA or GCA. The inhibitors inhibited sphere formation dramatically (k, l). Data show the mean ± s.e.m. Statistical significance was determined by Student’s t-test, * P < 0.05; **P < 0.01 (compared to DMSO). (m, n) BFA and FAO inhibitors reduce CSC in DU145, HT29, MCF7 and MDA-MB-231 cells. m, Flow cytometry was used to detect cancer stem cell surface makers in DU145, HT29 and MDA-MB-231 cells after 2 days of treatment with BFA. Cell subpopulations enriched with cancer stem cells (CD44+ and CD24- for DU145 and MDA-MB-231, CD44+ for HT29) were marked with red line. n, Crystal violet staining was used to detect cell survival after 2 days of treatment with triascin C or etomoxir using indicated concentrations in DU145, HT29 and MCF7 cells (left). Spheres were cultured with or without triascin C or etomoxir. The inhibitors markedly inhibited sphere formation in DU145 and MCF7 cells (right). Data show the mean ± s.e.m. Statistical significance was determined by Student’s t-test, **, P < 0.01 (compared to DMSO).
Supplementary information
A dying ISC is engulfed by a neighbouring EC
The genotype esgts>Arf79FRNAi - guts were stained with antibodies to Arm (red), GFP (green) and DAPI (blue). (WMV 270 kb)
Rights and permissions
About this article
Cite this article
Singh, S., Zeng, X., Zhao, J. et al. The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 538, 109–113 (2016). https://doi.org/10.1038/nature19788
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19788
This article is cited by
-
m6A demethylase FTO promotes tumor progression via regulation of lipid metabolism in esophageal cancer
Cell & Bioscience (2022)
-
Intestine-enriched apolipoprotein b orthologs are required for stem cell progeny differentiation and regeneration in planarians
Nature Communications (2022)
-
c-Myc-driven glycolysis polarizes functional regulatory B cells that trigger pathogenic inflammatory responses
Signal Transduction and Targeted Therapy (2022)
-
Neuronal accumulation of peroxidated lipids promotes demyelination and neurodegeneration through the activation of the microglial NLRP3 inflammasome
Nature Aging (2021)
-
Arf1-mediated lipid metabolism sustains cancer cells and its ablation induces anti-tumor immune responses in mice
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.