Abstract
Post-translational protein modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins such as small ubiquitin-like modifier (SUMO) regulates processes including protein homeostasis, the DNA damage response, and the cell cycle. Proliferating cell nuclear antigen (PCNA) is modified by Ub or poly-Ub at lysine (Lys)164 after DNA damage to recruit repair factors. Yeast PCNA is modified by SUMO on Lys164 and Lys127 during S-phase to recruit the anti-recombinogenic helicase Srs2. Lys164 modification requires specialized E2/E3 enzyme pairs for SUMO or Ub conjugation. For SUMO, Lys164 modification is strictly dependent on the E3 ligase Siz1, suggesting the E3 alters E2 specificity to promote Lys164 modification. The structural basis for substrate interactions in activated E3/E2–Ub/Ubl complexes remains unclear. Here we report an engineered E2 protein and cross-linking strategies that trap an E3/E2–Ubl/substrate complex for structure determination, illustrating how an E3 can bypass E2 specificity to force-feed a substrate lysine into the E2 active site.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006)
Gareau, J. R. & Lima, C. D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nature Rev. Mol. Cell Biol. 11, 861–871 (2010)
Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009)
Streich, F. C., Jr & Lima, C. D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 43, 357–379 (2014)
Sampson, D. A., Wang, M. & Matunis, M. J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 (2001)
Bernier-Villamor, V., Sampson, D. A., Matunis, M. J. & Lima, C. D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 (2002)
Reverter, D. & Lima, C. D. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435, 687–692 (2005)
Yunus, A. A. & Lima, C. D. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nature Struct. Mol. Biol . 13, 491–499 (2006)
Eddins, M. J., Carlile, C. M., Gomez, K. M., Pickart, C. M. & Wolberger, C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Struct. Mol. Biol . 13, 915–920 (2006)
Bosanac, I. et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 408, 420–431 (2011)
Wickliffe, K. E., Lorenz, S., Wemmer, D. E., Kuriyan, J. & Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 (2011)
Saha, A., Lewis, S., Kleiger, G., Kuhlman, B. & Deshaies, R. J. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 (2011)
Page, R. C., Pruneda, J. N., Amick, J., Klevit, R. E. & Misra, S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry 51, 4175–4187 (2012)
Rodrigo-Brenni, M. C., Foster, S. A. & Morgan, D. O. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559 (2010)
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009)
Plechanovová, A., Jaffray, E. G., Tatham, M. H., Naismith, J. H. & Hay, R. T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012)
Dou, H., Buetow, L., Sibbet, G. J., Cameron, K. & Huang, D. T. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature Struct. Mol. Biol . 19, 876–883 (2012)
Pruneda, J. N. et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012)
Dou, H., Buetow, L., Sibbet, G. J., Cameron, K. & Huang, D. T. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nature Struct. Mol. Biol . 20, 982–986 (2013)
Scott, D. C. et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 (2014)
Buetow, L. et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol. Cell 58, 297–310 (2015)
Wright, J. D., Mace, P. D. & Day, C. L. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nature Struct. Mol. Biol . 23, 45–52 (2016)
Rytinki, M. M., Kaikkonen, S., Pehkonen, P., Jääskeläinen, T. & Palvimo, J. J. PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66, 3029–3041 (2009)
Johnson, E. S. & Gupta, A. A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 (2001)
Yunus, A. A. & Lima, C. D. Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 (2009)
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002)
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007)
Parker, J. L. & Ulrich, H. D. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J . 28, 3657–3666 (2009)
Pfander, B., Moldovan, G. L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005)
Papouli, E. et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 (2005)
Armstrong, A. A., Mohideen, F. & Lima, C. D. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 (2012)
Parker, J. L. & Ulrich, H. D. A SUMO-interacting motif activates budding yeast ubiquitin ligase Rad18 towards SUMO-modified PCNA. Nucleic Acids Res . 40, 11380–11388 (2012)
Mascle, X. H. et al. Identification of a non-covalent ternary complex formed by PIAS1, SUMO1, and UBC9 proteins involved in transcriptional regulation. J. Biol. Chem. 288, 36312–36327 (2013)
Cappadocia, L., Pichler, A. & Lima, C. D. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nature Struct. Mol. Biol . 22, 968–975 (2015)
Knipscheer, P., van Dijk, W. J., Olsen, J. V., Mann, M. & Sixma, T. K. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J . 26, 2797–2807 (2007)
Capili, A. D. & Lima, C. D. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 369, 608–618 (2007)
Duda, D. M. et al. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J. Mol. Biol. 369, 619–630 (2007)
Stehmeier, P. & Muller, S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol. Cell 33, 400–409 (2009)
Jentsch, S. & Psakhye, I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 47, 167–186 (2013)
Chang, L., Zhang, Z., Yang, J., McLaughlin, S. H. & Barford, D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522, 450–454 (2015)
McGinty, R. K., Henrici, R. C. & Tan, S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014)
Mattiroli, F., Uckelmann, M., Sahtoe, D. D., van Dijk, W. J. & Sixma, T. K. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nature Commun . 5, 3291 (2014)
Parker, J. L. et al. SUMO modification of PCNA is controlled by DNA. EMBO J . 27, 2422–2431 (2008)
Lois, L. M. & Lima, C. D. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J . 24, 439–451 (2005)
Yunus, A. A. & Lima, C. D. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol. Biol. 497, 167–186 (2009)
Knipscheer, P. et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol. Cell 31, 371–382 (2008)
Mossessova, E. & Lima, C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000)
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (eds Carter Jr, C. W. & Sweet, R. M. ) 307–326 (Academic, 1997)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Acknowledgements
Research at NE-CAT beamlines was funded by P41 GM103403 (National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS)), S10 RR029205 (NIH-ORIP (Office of Research Infrastructure Programs) High-End Shared Instrument grant) at the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (DE-AC02-06CH11357). Research was supported in part by GM065872 and GM118080 (NIH/NIGMS, C.D.L.) and P30CA008748 (NIH/ National Cancer Institute). The content is the authors’ responsibility and does not represent the official views of the NIH. C.D.L. is a Howard Hughes Medical Institute Investigator.
Author information
Authors and Affiliations
Contributions
F.C.S. and C.D.L. executed experiments, data analysis, and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks B. Eichman, R. Hay and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
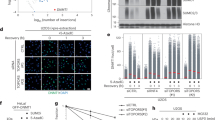
Extended Data Figure 1 E2Ubc9–SUMO thioester mimic and cross-linking to substrate PCNA for reconstitution with E3Siz1.
a, SDS–PAGE analysis of in vitro  or
or  charging with SUMO in the presence and absence of
charging with SUMO in the presence and absence of  at pH (7.5) (left) and purification of the
at pH (7.5) (left) and purification of the  –SUMO (middle) and
–SUMO (middle) and  –SUMO (right) thioester mimetics. b, Rates for in vitro SUMO modification of PCNA in assays using various concentrations of purified E2Ubc9–SUMOD68R–Alexa488-labelled thioester, 1 nM
–SUMO (right) thioester mimetics. b, Rates for in vitro SUMO modification of PCNA in assays using various concentrations of purified E2Ubc9–SUMOD68R–Alexa488-labelled thioester, 1 nM  , and 32 μM PCNA with 0, 2, 5, or 20 μM of the
, and 32 μM PCNA with 0, 2, 5, or 20 μM of the  –SUMO or
–SUMO or  –SUMO thioester mimic (left) with exemplary non-reducing SDS–PAGE for the 0.5 μM E2Ubc9–SUMOD68R–Alexa488 reactions (right). The calculated Km and Ki from these fits are shown in Extended Data Tables 1 and 3 and the quantified data show mean ± s.d. (n = 3 technical replicates). c, SDS–PAGE analysis (left) of numbered 0.5 ml fractions from Superose6 analytical gel-filtration analysis (right) of complex reconstitution between E2Ubc9–SUMO–BMOE–PCNA and
–SUMO thioester mimic (left) with exemplary non-reducing SDS–PAGE for the 0.5 μM E2Ubc9–SUMOD68R–Alexa488 reactions (right). The calculated Km and Ki from these fits are shown in Extended Data Tables 1 and 3 and the quantified data show mean ± s.d. (n = 3 technical replicates). c, SDS–PAGE analysis (left) of numbered 0.5 ml fractions from Superose6 analytical gel-filtration analysis (right) of complex reconstitution between E2Ubc9–SUMO–BMOE–PCNA and  (green) or the
(green) or the  –SUMO fusion (blue). Elution profiles for E2Ubc9–SUMO–BMOE–PCNA (purple) and
–SUMO fusion (blue). Elution profiles for E2Ubc9–SUMO–BMOE–PCNA (purple) and  (red) alone are shown. d, The normalized change in polarization observed upon addition of serially diluted E2Ubc9 with Alexa488 labelled SUMO or SUMOD68R. Data were fitted to a single-site binding model accounting for receptor depletion. Data show mean ± s.d. (n = 3 technical replicates). For gel source data, see Supplementary Fig. 1.
(red) alone are shown. d, The normalized change in polarization observed upon addition of serially diluted E2Ubc9 with Alexa488 labelled SUMO or SUMOD68R. Data were fitted to a single-site binding model accounting for receptor depletion. Data show mean ± s.d. (n = 3 technical replicates). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 2 Comparing strategies for crosslinking the E2–SUMO thioester mimic and substrate PCNA.
a, Chemical structures of the proposed tetrahedral intermediate formed during PCNA Lys164 attack of E2Ubc9–SUMO thioester (left), a BMOE cross-link (middle) or an EDT cross-link (right) between E2Ubc9–SUMO C93 and PCNA K164C. Indicated distances were estimated with ChemDraw15 (PerkinElmer). b, Control non-reducing SDS–PAGE panel for Fig. 1a showing mock-treated PCNA K127R/K164C (DMSO instead of EDT in DMSO) is unable to accept transthioesterification of SUMO at position 164. c, SDS–PAGE analysis of the 5 ml fractions from the final preparative Superdex200 gel-filtration purification of the E2Ubc9–SUMO–EDT–PCNA/ –SUMO complex. For gel source data, see Supplementary Fig. 1.
–SUMO complex. For gel source data, see Supplementary Fig. 1.
Extended Data Figure 3 E2Ubc9 active site, conformation of SUMOD, and comparison with relevant structures.
a, Stereo image of simulated annealing electron density map showing the EDT linkage and the SUMO Gly98 linkage to E2Ubc9 A129K. The 2Fo − Fc electron density map is contoured at 0.8σ (grey mesh). b, Alignment of the E2 enzymes from the current structure, SUMO-modified RanGAP1 bound to  and E3Znf451 (5D2M),
and E3Znf451 (5D2M),  -Ub bound to the RING dimer from E3BIRC7 (4AUQ), and E2
-Ub bound to the RING dimer from E3BIRC7 (4AUQ), and E2  -Ub bound to the RING dimer from E3RNF4 (4AP4) showing two orientations of the E2 active site. c, Model of tetrahedral intermediate generated by comparing our structure with other structures of E2–Ubl/E3 complexes, particularly Protein Data Bank (PDB) accession numbers 5DM2 and 4P5O. d, Alignment of the current structure and three E2/RING (PDB 1UR6, 3EB6, and 3FN1) complexes and one E2/UBox (PDB 2C2V) complex (aligned by the E2). e, Alignments of four E2–Ubl/E3 complexes (aligned by the E2) in the closed activated confirmation for the current structure,
-Ub bound to the RING dimer from E3RNF4 (4AP4) showing two orientations of the E2 active site. c, Model of tetrahedral intermediate generated by comparing our structure with other structures of E2–Ubl/E3 complexes, particularly Protein Data Bank (PDB) accession numbers 5DM2 and 4P5O. d, Alignment of the current structure and three E2/RING (PDB 1UR6, 3EB6, and 3FN1) complexes and one E2/UBox (PDB 2C2V) complex (aligned by the E2). e, Alignments of four E2–Ubl/E3 complexes (aligned by the E2) in the closed activated confirmation for the current structure,  –SUMO (PDB 5D2M),
–SUMO (PDB 5D2M),  -Ub (PDB 4AP4), and
-Ub (PDB 4AP4), and  -Nedd8 (PDB 4P5O). f, SDS–PAGE analysis of multiple turnover assays of SUMO modification of PCNA using in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 μM PCNA for the quantified data shown in Fig. 2c. g, Alignments of E2 from relevant structures with lysine or arginine residues within or projecting towards the E2 active sites compared with the current structure. Lysine 63 from acceptor ubiquitin projecting towards the active site of the
-Nedd8 (PDB 4P5O). f, SDS–PAGE analysis of multiple turnover assays of SUMO modification of PCNA using in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 μM PCNA for the quantified data shown in Fig. 2c. g, Alignments of E2 from relevant structures with lysine or arginine residues within or projecting towards the E2 active sites compared with the current structure. Lysine 63 from acceptor ubiquitin projecting towards the active site of the  –Ub is shown in green (PDB 2GMI). Lysine 524 from SUMO-modified RanGAP1 laying across the active site of
–Ub is shown in green (PDB 2GMI). Lysine 524 from SUMO-modified RanGAP1 laying across the active site of  is shown in magenta. The Lys720Arg from Cullin-1 projecting into the active site of E2Ubc12–Nedd8 is shown in grey (PDB 4P5O). For the current structure, EDT was removed from the model, Cys164 was mutated back to lysine, and the side chain was fitted to the electron density and is shown in pink in reference to the current E2 (blue) and donor SUMO (orange). For gel source data, see Supplementary Fig. 1.
is shown in magenta. The Lys720Arg from Cullin-1 projecting into the active site of E2Ubc12–Nedd8 is shown in grey (PDB 4P5O). For the current structure, EDT was removed from the model, Cys164 was mutated back to lysine, and the side chain was fitted to the electron density and is shown in pink in reference to the current E2 (blue) and donor SUMO (orange). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 4 SUMOB bound to the E2 backside enhances E2Ubc9–SUMO recruitment.
a, Alignment of the current E2Ubc9/backside SUMOB (left) to previously observed E2Ubc9/backside SUMO complexes (right). The position of the D68R mutation is shown in red sticks (left). b, Primary E3Siz1 structure (top). Cartoons indicating the E3Siz1 or E3Siz1–SUMO fusion constructs used in the multiple turnover in vitro assays (middle) shown in Fig. 3 using a titration of the purified E2Ubc9–SUMOD68R–Alexa488 thioester with or without 1.5-fold excess of the indicated additional molecule of non-conjugatable SUMO, 1 nM of the indicated E3 construct, and 32 μM PCNA. Representative non-reducing SDS–PAGE showing the 0.5 μM E2Ubc9–SUMOD68R–Alexa488 thioester reactions below the plots of the rates of reaction for each E2Ubc9–SUMOD68R concentration (middle). The kinetics of SUMO modification of PCNA were calculated and Km and kcat determined (bottom); these are shown in Extended Data Table 3. The quantified rate data show mean ± s.d. (n = 3 technical replicates). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 5 E2Ubc9 and E3Siz1 determinants of lysine specificity.
a, Plots of the rates observed at different pH values for multiple turnover in vitro assays of SUMO modification of PCNA using 0.1 μM purified E2Ubc9–SUMOD68R–Alexa488 thioester (or E2Ubc9 mutant thioesters) with 5 nM E3Siz1 and 4 μM PCNA at 4 °C. b, SDS–PAGE analysis of multiple turnover assays of SUMO modification of PCNA using in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 μM PCNA for the quantified data shown in Fig. 4d. c, SDS–PAGE analysis of multiple turnover assays of SUMO modification of PCNA using in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 μM PCNA and quantified. d, Representative non-reducing SDS–PAGE analysis of the single turnover in vitro assays of SUMO modification of PCNA shown in Fig. 4e. These assays utilized 5 nM of the E2Ubc9–SUMOD68R–Alexa488 thioester (or E2Ubc9 mutant thioesters) in reactions with 50 nM of the indicated E3Siz1 and a titration of PCNA. Shown are typical SDS–PAGE analyses from the 10 μM PCNA reactions. The data were used to extract the kinetic constants for the reactions shown as histograms and in Extended Data Table 5. For a, c, and d the quantified rate data show mean ± s.d. (n = 3 technical replicates). For gel source data, see Supplementary Fig. 1.
Extended Data Figure 6 Shape complementarity between the E2Ubc9–SUMO/E3 complex and PCNA.
a, The current structure (colour) with the crystallographic packing of a lattice mate PCNA molecule (black). b, Non-reducing SDS–PAGE analysis of 2 min endpoint in vitro E2Ubc9–SUMO thioester formation reactions with 0.05 μM E1, 0.4 μM of the indicated E2Ubc9, and 22 μM SUMO (left) and the quantified E2–SUMO band (right). The quantified band intensity shows mean ± s.d. (n = 3 technical replicates). c, SDS–PAGE analysis of multiple turnover assays of SUMO modification of PCNA using in vitro reactions with coupled E1 (200 nM), E2 (100 nM), and E3 (50 nM) activities with 4 μM PCNA or without PCNA (diSUMO formation) shown quantified in Fig. 5c. d, Location of E2Ubc9 and PCNA mutations that had no effect (red sticks) on activities observed for in vitro assays similar to those performed in Fig. 5c in relation to residues that did show effects (green sticks). e, The  –Ub was aligned to E2Ubc9 in the current structure and subsequently the Lys164/Glu165 loop from trimeric PCNA (pink) was aligned onto the Lys63/Glu64 loop from acceptor ubiquitin (PDB 2GMI, green). Within this conformation the E3Siz1 PINIT domain (cyan) clashes with another protomer of the PCNA trimer (grey). For gel source data, see Supplementary Fig. 1.
–Ub was aligned to E2Ubc9 in the current structure and subsequently the Lys164/Glu165 loop from trimeric PCNA (pink) was aligned onto the Lys63/Glu64 loop from acceptor ubiquitin (PDB 2GMI, green). Within this conformation the E3Siz1 PINIT domain (cyan) clashes with another protomer of the PCNA trimer (grey). For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Figures
This file contains gel source data for Figure 1 and Extended Data Figures 1-6. (PDF 1169 kb)
Rights and permissions
About this article
Cite this article
Streich Jr, F., Lima, C. Capturing a substrate in an activated RING E3/E2–SUMO complex. Nature 536, 304–308 (2016). https://doi.org/10.1038/nature19071
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19071
This article is cited by
-
The study on interacting factors and functions of GASA6 in Jatropha curcas L.
BMC Plant Biology (2023)
-
A bifunctional molecule-assisted synthesis of mimics for use in probing the ubiquitination system
Nature Protocols (2023)
-
Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer
Nature Communications (2023)
-
SUMOylation of RNF146 results in Axin degradation and activation of Wnt/β-catenin signaling to promote the progression of hepatocellular carcinoma
Oncogene (2023)
-
Signalling mechanisms and cellular functions of SUMO
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.