Abstract
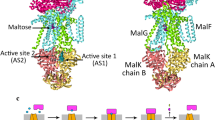
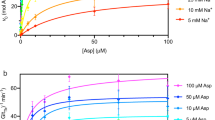
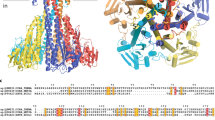
Transport of solutes across biological membranes is performed by specialized secondary transport proteins in the lipid bilayer1, and is essential for life. Here we report the structures of the sodium-independent carnitine/butyrobetaine antiporter CaiT from Proteus mirabilis (PmCaiT) at 2.3-Å and from Escherichia coli (EcCaiT) at 3.5-Å resolution. CaiT belongs to the family of betaine/carnitine/choline transporters (BCCT), which are mostly Na+ or H+ dependent, whereas EcCaiT is Na+ and H+ independent2. The three-dimensional architecture of CaiT resembles that of the Na+-dependent transporters LeuT3 and BetP4, but in CaiT a methionine sulphur takes the place of the Na+ ion to coordinate the substrate in the central transport site, accounting for Na+-independent transport. Both CaiT structures show the fully open, inward-facing conformation, and thus complete the set of functional states that describe the alternating access mechanism5. EcCaiT contains two bound butyrobetaine substrate molecules, one in the central transport site, the other in an extracellular binding pocket. In the structure of PmCaiT, a tryptophan side chain occupies the transport site, and access to the extracellular site is blocked. Binding of both substrates to CaiT reconstituted into proteoliposomes is cooperative, with Hill coefficients up to 1.7, indicating that the extracellular site is regulatory. We propose a mechanism whereby the occupied regulatory site increases the binding affinity of the transport site and initiates substrate translocation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Coordinates and structure factors for P. mirabilis CaiT and E. coli CaiT with bound substrate are deposited in the Protein Data Bank under accession numbers 2WSW and 2WSX, respectively..
References
Csaky, T. Z. Transport through biological membranes. Annu. Rev. Physiol. 27, 415–450 (1965)
Jung, H. et al. CaiT of Escherichia coli, a new transporter catalyzing l-carnitine/γ-butyrobetaine exchange. J. Biol. Chem. 277, 39251–39258 (2002)
Yamashita, A., Singh, S. K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homologue of Na+/Cl–-dependent neurotransmitter transporters. Nature 437, 215–223 (2005)
Ressl, S., Terwisscha van Scheltinga, A. C., Vonrhein, C., Ott, V. & Ziegler, C. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52 (2009)
Diallinas, G. Biochemistry. An almost-complete movie. Science 322, 1644–1645 (2008)
Vinothkumar, K. R., Raunser, S., Jung, H. & Kühlbrandt, W. Oligomeric structure of the carnitine transporter CaiT from Escherichia coli . J Biol Chem 281, 4795–4801 (2006)
Horn, C. et al. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357, 592–606 (2006)
Padgett, C. L., Hanek, A. P., Lester, H. A., Dougherty, D. A. & Lummis, S. C. Unnatural amino acid mutagenesis of the GABAA receptor binding site residues reveals a novel cation–pi interaction between GABA and β2Tyr97. J. Neurosci. 27, 886–892 (2007)
Schiefner, A. et al. Cation–pi interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli . J. Biol. Chem. 279, 5588–5596 (2004)
Torrice, M. M., Bower, K. S., Lester, H. A. & Dougherty, D. A. Probing the role of the cation–pi interaction in the binding sites of GPCRs using unnatural amino acids. Proc. Natl Acad. Sci. USA 106, 11919–11924 (2009)
Shaffer, P. L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+ independent amino acid transporter. Science 325, 1010–1014 (2009)
Shi, L., Quick, M., Zhao, Y., Weinstein, H. & Javitch, J. A. The mechanism of a neurotransmitter: sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol. Cell 30, 667–677 (2008)
Farwick, M., Siewe, R. M. & Kramer, R. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum . J. Bacteriol. 177, 4690–4695 (1995)
Weyand, S. et al. Structure and molecular mechanism of a nucleobase–cation–symport-1 family transporter. Science 322, 709–713 (2008)
Ricard, J. & Cornish-Bowden, A. Co-operative and allosteric enzymes: 20 years on. Eur. J. Biochem. 166, 255–272 (1987)
Barros, T., Royant, A., Standfuss, J., Dreuw, A. & Kuhlbrandt, W. Crystal structure of plant light-harvesting complex shows the active, energy-transmitting state. EMBO J. 28, 298–306 (2009)
Faham, S. et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 (2008)
Gao, X. et al. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 (2009)
Sevilla, A. et al. Design of metabolic engineering strategies for maximizing l-(−)-carnitine production by Escherichia coli. Integration of the metabolic and bioreactor levels. Biotechnol. Prog. 21, 329–337 (2005)
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460, 1040–1043 (2009)
Tang, L., Bai, L., Wang, W. H. & Jiang, T. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nature Struct. Mol. Biol. 17, 492–496 (2010)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
The. CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Rossmann, M. G. & Blow, D. M. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 15, 24–31 (1962)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Terwilliger, T. C. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr. D 60, 2144–2149 (2004)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Fenn, T. D., Ringe, D. & Petsko, G. A. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Cryst. 36, 944–947 (2003)
DeLano, W. L. The PyMOL molecular graphics system. (DeLano Scientific, San Carlos, CA, USA). (2002)
Krissinel, E. & Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004)
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
Holm, L. & Sander, C. Mapping the protein universe. Science 273, 595–603 (1996)
Acknowledgements
We thank Ö. Yildiz, T. Barros and R. Wouts for help with computing; S. Ressl for providing the BetP model; K. R Vinothkumar for growing the first crystals of CaiT, J. Hakulinen; F. Joos for help with transport measurements; H. Jung and K. Fendler for discussions; and C. Ziegler and L. Forrest for reading the manuscript. The γ-butyrobetaine used for binding studies was a gift from Lonza (Switzerland). We also thank A. Pauluhn and the X10SA beamline staff at the Swiss Light Source, and the beamline staff at the European Synchrotron Radiation Facility.
Author information
Authors and Affiliations
Contributions
Experiments were performed by S.S. and U.G.; cloning and mutagenesis was performed by S.K. Crystals were grown by S.S., diffraction data were collected and processed by S.S. and A.C.T.v.S., and the structures were analysed by S.S., A.C.T.v.S. and W.K. S.S. and W.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Background Notes, Supplementary Tables 1- 4, Supplementary Figures 1- 9 with legends and additional references. (PDF 22457 kb)
Rights and permissions
About this article
Cite this article
Schulze, S., Köster, S., Geldmacher, U. et al. Structural basis of Na+-independent and cooperative substrate/product antiport in CaiT. Nature 467, 233–236 (2010). https://doi.org/10.1038/nature09310
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09310
This article is cited by
-
Conformational transition induced in the aspartate:alanine antiporter by l-Ala binding
Scientific Reports (2022)
-
Comparison of the functional properties of trimeric and monomeric CaiT of Escherichia coli
Scientific Reports (2019)
-
Locking Two Rigid-body Bundles in an Outward-Facing Conformation: The Ion-coupling Mechanism in a LeuT-fold Transporter
Scientific Reports (2019)
-
Monoamine transporters: structure, intrinsic dynamics and allosteric regulation
Nature Structural & Molecular Biology (2019)
-
Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state
Nature Structural & Molecular Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.