Abstract
Genomes are organized into high-level three-dimensional structures, and DNA elements separated by long genomic distances can in principle interact functionally. Many transcription factors bind to regulatory DNA elements distant from gene promoters. Although distal binding sites have been shown to regulate transcription by long-range chromatin interactions at a few loci, chromatin interactions and their impact on transcription regulation have not been investigated in a genome-wide manner. Here we describe the development of a new strategy, chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) for the de novo detection of global chromatin interactions, with which we have comprehensively mapped the chromatin interaction network bound by oestrogen receptor α (ER-α) in the human genome. We found that most high-confidence remote ER-α-binding sites are anchored at gene promoters through long-range chromatin interactions, suggesting that ER-α functions by extensive chromatin looping to bring genes together for coordinated transcriptional regulation. We propose that chromatin interactions constitute a primary mechanism for regulating transcription in mammalian genomes.
Similar content being viewed by others
Main
Although genomic information is usually presented as a linear series of bases, genomes are known to be organized into three-dimensional structures in vivo through interactions with protein factors for nuclear process such as transcription1. The precise and coordinated regulation of transcription requires the binding of transcription factors to specific regulatory DNA sequences in the genome. Chromatin immunoprecipitation (ChIP) microarray2 (ChIP-Chip) and ChIP-sequencing3,4 (ChIP-PET and ChIP-Seq) have identified global transcription-factor-binding sites (TFBSs) and revealed that many TFBSs are far from gene promoters5. For example, most TFBSs bound by ER-α in the human genome are distal to transcription start sites (TSSs) of target genes6,7,8,9,10. A major question arising from this observation is which distal TFBSs are non-functional fortuitous binding sites, and which are involved in transcriptional activity through a remote control mechanism. Long-range chromatin interactions between DNA elements engaged in transcriptional regulation11,12 have been observed with the use of chromosome conformation capture (3C)13,14 and variants including ChIP-3C15,16, 4C17,18,19,20, 5C21 and 6C22, and also RNA tagging and recovery of associated proteins (RNA TRAP)23 and fluorescence in situ hybridization (FISH)24. However, these methods are limited to one-point or partial genome-oriented detection and are incapable of the de novo detection of genome-wide chromatin interactions25.
To address whether and how DNA elements bound by protein factors interact through long-range chromatin looping in a genome-wide and unbiased manner, we conceived a new strategy, which we called ChIA-PET. We applied ChIA-PET to characterize ER-α-bound chromatin interactions in oestrogen-treated human breast adenocarcinoma cells (MCF-7), and generated the first human chromatin interactome map. (A ChIA-PET visualization browser is provided at http://cms1.gis.a-star.edu.sg (username ‘guest’, password ‘gisimsgtb’) for viewing the ER-α ChIA-PET map.) Furthermore, using active promoter and transcriptional marks such as trimethylation of lysine 4 on histone H3 (H3K4me3) and RNA polymerase II (RNAPII) from ChIP sequencing as well as gene expression microarray data, we show that ER-α-bound chromatin interactions are functionally involved in regulating specific genes.
The ChIA-PET method
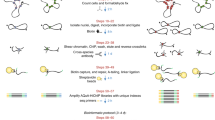
In ChIA-PET, long-range chromatin interactions are captured by crosslinking with formaldehyde. Sonicated DNA–protein complexes are enriched by ChIP. Tethered DNA fragments in each of the chromatin complexes are connected with DNA linkers by proximity ligation, and paired-end tags (PETs) are extracted for sequencing. The resulting ChIA-PET sequences are mapped to reference genomes to reveal relationships between remote chromosomal regions brought together into close spatial proximity by protein factors (Fig. 1a and Supplementary Fig. 1).
a, ChIA-PET diagram. DNA fragments in sonicated, ChIP-enriched chromatin complexes were processed by linker ligation, proximity ligation, PET extraction, sequencing, and mapping to reveal interacting loci. b, ChIA-PET browser tracks: 1, H3K4me3 ChIP-Seq; 2, RNAPII ChIP-Seq; 3, ER-α (orange) and FoxA1 ChIP-chip (green)9; 4, ER-α ChIA-PET density; 5, inter-ligation PETs. Inset: 3C validation of interacting ER-αBSs (purple) and controls (blue) under ethanol control (ET) and induction with oestrogen (E2). Error bars show s.e.m. c, 4C validation, showing 4C bait region (blue) and interaction targets (purple bars) with counts of sequence hits. d, Validation by FISH, showing increased P2–P1 interactions under E2 induction with background normalization (P3/P2). FISH probe genomic locations (P1, P2 and P3) are indicated.
ChIA-PET proximity ligation generates two types of ligation products: self-ligation of the same DNA fragments and inter-ligation between different DNA fragments. PET sequences derived from self-ligation products are mapped in the reference genome within a 3-kilobase (kb) span, demarcating ChIP DNA fragments, similar to the standard ChIP-sequencing method3,8. Tethered DNA fragments in individual chromatin complexes can also ligate with each other, and the mapping results of such inter-ligation PET sequences would reveal if they are intrachromosomal (both tags of each PET are from the same chromosome) or interchromosomal (the tags are from different chromosomes). Singleton PETs are presumed experimental noise, and overlapping PET clusters are considered enriched putative binding sites or interaction events (Supplementary Fig. 2).
To test the ChIA-PET strategy, we constructed two ChIA-PET libraries from independent ER-α ChIP-enriched oestrogen-treated MCF-7 chromatin preparations, and generated two replicate pilot datasets (IHM001H and IHM001N) using Roche/454 pyrosequencing. Our analysis showed that both ChIA-PET libraries produced comparable putative binding sites and interactions. To assess levels of false positive chromatin interactions, we created a negative control ChIP-PET library (IHM043) from the same ChIP sample, wherein the DNA was reverse crosslinked before proximity ligation. We also analysed a previously reported cloning-based ChIP-PET library (SHC007)8. Both libraries generated abundant binding sites but no interactions. As an additional control, we used IgG, which binds to chromatin nonspecifically, to perform a mock ChIA-PET analysis (IHM062), and only a few binding sites and interactions were identified (Table 1, Supplementary Figs 2 and 3 and Supplementary Note 1).
In proximity ligation-based analyses including 3C, the level of non-specific chimaeric DNA ligations between different chromatin complexes can be high and thus may confound data analysis. To address this, we designed linker nucleotide barcodes in the ChIA-PET method to specifically identify such chimaeric ligation PETs in another ER-α ChIA-PET replicate. Linker barcoding analysis suggests that chimaeric ligations are random and do not overlap with each other to form false positive interactions (Table 1, Supplementary Fig. 4 and Supplementary Note 2). A possible complication is that ChIP-enriched loci with more DNA fragments would result in proportionally higher chances of inter-ligations, leading to false-positive interactions comprising randomly overlapping inter-ligation PETs among highly-enriched ChIP DNA fragments. Hence, we devised a statistical scheme to calculate such probabilities and neutralize the potential ChIP-enrichment bias (Supplementary Methods; validations are given in Supplementary Fig. 5).
Together, these libraries indicate that the prevalent chromatin interactions (Supplementary Fig. 2d–g) identified by ER-α ChIA-PET data depend on proximity ligations of chromatin complexes and are neither technical artefacts of ligations between random DNA fragments nor mapping errors.
ER-α-bound chromatin interactome map
Next we generated a large ER-α ChIA-PET dataset (IHM001F) with 3.2 × 107 PET sequences by Illumina GAII paired-end sequencing (Table 1 and Supplementary Methods) for comprehensive analyses of ER-α binding and chromatin interactions in oestrogen-treated MCF-7 cells. Of 4.6 × 106 uniquely mapped PET sequences, 1.2 × 106 (27%) were self-ligation PETs. Among the self-ligation PETs, 16.7% clustered to form overlapping PET groups, representing 14,468 putative ER-α-binding sites (ER-αBSs) (false discovery rate (FDR) < 0.01, PET count per ER-αBS at least 5; Supplementary Table 1). Of the inter-ligation PETs, 2.3 × 105 (5.1% of uniquely aligned PETs) were intrachromosomal and 3.2 × 106 (68%) were interchromosomal (Table 1). After statistical analyses in which we discarded singleton inter-ligation PETs as either very weak interactions or background noise, defined clusters from overlapping inter-ligation PETs, corrected for ChIP enrichment biases and filtered out obviously false interactions due to structural variations in the MCF-7 genome (Supplementary Methods), we identified a large set of 1,451 intrachromosomal and a small set of 15 interchromosomal overlapping clusters consisting of three or more inter-ligation PETs per cluster (FDR < 0.05). These represent paired inter-ligating ChIP DNA fragments, which indicate potential distant chromatin interactions bound by ER-α (Supplementary Table 2).
Each chromatin interaction detected by an inter-ligation PET cluster features two anchor regions (interacting loci) and a loop (the intermediate genomic span between the two anchors), and is therefore called a ‘duplex interaction’ (Supplementary Table 2). Most anchors (1,893 out of 2,008 = 94%) involve self-ligation PET-defined ER-αBSs (FDR < 0.01). Many nearby duplex interactions are interconnected, linking three or more anchors into ‘daisy-chain’ aggregated complex interactions (Fig. 1b–d and Supplementary Fig. 6). For example, multiple duplex interactions with three ER-αBSs in the SIAH2 region interconnect to form a complex interaction. Hence, we further assembled 1,036 duplex interactions into 274 complex interactions based on overlapping of interaction anchors (Supplementary Methods). The remaining interactions (415) were stand-alone duplex interactions. In all, we identified 689 ER-α-bound chromatin interaction regions (Supplementary Table 3).
To verify the ChIA-PET results, we validated several new ER-αBSs identified in this study by ChIP-quantitative polymerase chain reaction (qPCR) (Supplementary Fig. 7), as well as putative intrachromosomal interaction sites (20 genomic loci) by 3C, ChIP-3C, 4C and FISH experiments (three examples are shown in Fig. 1; others are shown in Supplementary Figs 8–11 and Supplementary Tables 4 and 5). Moreover, the 3C and FISH experiments showed higher levels of chromatin interactions in oestrogen-treated conditions than in untreated conditions, indicating that the interactions are oestrogen-dependent. We also examined three putative interchromosomal interactions by FISH; however, none of them were positive (Supplementary Table 4 and Supplementary Note 3), suggesting that most ER-α-bound intrachromosomal interactions were genuine, whereas the putative interchromosomal interactions were false positives or were too weak to be validated.
Taken together, the ER-αBS and chromatin interactions identified by ChIA-PET data constitute a whole-genome chromatin interaction map bound by ER-α. The genomic spans of most duplex interactions (86%) are less than 100 kb, about 13% are from 100 kb to 1 megabase (Mb), and less than 1% are more than 1 Mb. Complex interactions extend genomic span by connecting multiple duplex interactions. Many complex interactions (47%) have genomic spans in the range 100 kb to 1 Mb, with a few that are more than 1 Mb (Supplementary Fig. 12 and Supplementary Table 3).
To determine the reproducibility of this chromatin interactome map, we generated an additional ER-α ChIA-PET library by using a different antibody against ER-α10. For this biological replicate (IHH015F), we obtained 2.0 × 107 PET sequences (Table 1 and Supplementary Methods). Overall, the two ER-α ChIA-PET libraries were very similar, with many overlapping ER-αBSs and intrachromosomal interactions but few interchromosomal interactions (Table 1 and Supplementary Tables 1 and 2). The ER-αBSs identified in these two libraries showed high reproducibility, especially for highly enriched binding peaks. The 2,513 ER-αBSs with at least 50 PET counts per cluster (high enrichment) overlapped with more than 70% of the ER-αBSs in the replicate ChIA-PET library (Supplementary Table 6). Furthermore, these high-enrichment ER-αBSs intersected well with previously reported ER-α binding maps9,10 (Fig. 2a and Supplementary Fig. 13). High-enrichment ER-αBSs are therefore more reliable than low-enrichment sites. Many intrachromosomal interaction regions are detected in both replicate libraries. Highly abundant chromatin interactions are mostly reproducible. Of the top 100 most abundant chromatin interactions in IHM001F, 86 were found in IHH015F (more analyses are given in Supplementary Table 7). Furthermore, all interactions previously identified and validated in this study are found in both replicate libraries (Supplementary Table 5). Conversely, none of the putative interchromosomal interactions were reproducible.
a, Numbers of ER-αBSs identified with different ChIP enrichment cutoffs and reproducibility analyses as measured by overlapping with another ChIA-PET dataset (IHH015F), ChIP-Seq10 and ChIP-chip7 data. Left: low-enrichment ER-αBSs, 11,955 (5–49 PETs per site). Right: high-enrichment ER-αBSs, 2,513 (50 or more PETs per site). b–d, Examples of ER-αBSs involved in complex interactions (b), duplex interactions (c) and no interactions (d) (singleton inter-ligation PETs only (right) or no inter-ligation PETs (left)). e, ER-αBS distribution in different categories of interactions as exemplified in b–d. Left: low-enrichment ER-αBSs. Right: high-enrichment ER-αBSs.
Taken together, our results demonstrate that the ChIA-PET method is highly reliable. Furthermore, our data suggest that ER-α functions primarily by means of an intrachromosomal mechanism. Our subsequent analyses therefore focused on intrachromosomal interactions. Downstream analyses for both ChIA-PET replicate libraries showed similar results; for simplicity, we discuss our results here using IHM001F, but results for IHH015F are given in Supplementary Note 4.
We examined how many ER-αBSs are involved in complex and duplex interactions, or in no interactions (Fig. 2b–d). Our analysis showed that high-enrichment ER-αBSs are much more frequently involved in interactions (53%) than are low-enrichment ER-αBSs (only 9%) (Fig. 2e and Supplementary Fig. 13), suggesting that high-confidence and strong ER-αBSs are more likely than weaker ER-αBSs to be involved in chromatin interactions. To gain a better understanding of ER-αBSs with respect to ER-α target genes, we analysed how many ER-αBSs are proximal or distal to gene promoters, based on a cutoff of 5 kb from transcription start sites (TSSs) of University of California Santa Cruz (UCSC) gene database26. Of 2,342 ER-αBSs involved in chromatin interactions, 387 (17%) were proximal and 1,955 (83%) were distal to TSSs (Supplementary Fig. 14). We also observed the same ratio for non-interacting ER-αBSs: 2,043 (17%) were proximal and 10,175 (83%) were distal. Most ER-αBSs are therefore distal to gene TSSs, which is in agreement with previous studies7,8,10.
Chromatin interaction and transcriptional regulation
To investigate the functions of ER-αBSs and ER-α-bound chromatin interactions in transcription activation, we generated genome-wide maps of H3K4me3 and RNAPII ChIP-Seq data from MCF-7 cells under oestrogen induction (Supplementary Methods). H3K4me3 is a histone modification that specifically marks active promoters27, and the presence of RNAPII is strong evidence for genes that are actively transcribing28. We also analysed previously reported FoxA1 ChIP-Chip data9, because FoxA1 is an important cofactor of ER-α6,9. Generally, H3K4me3, RNAPII and FoxA1 marks showed enrichment around ER-αBSs in our analyses (Fig. 3a). When we compared interacting ER-αBSs with non-interacting ER-αBSs, we found a significant enrichment gradient of RNAPII and FoxA1 binding around ER-αBSs: most association was with ER-αBSs involved in complex interactions, followed by duplex interactions, and lastly no interactions (Fig. 3a and Supplementary Fig. 15a; significance tests are shown in Supplementary Note 5).
a, Association of ER-αBSs in complex-interaction (left), duplex-interaction (middle) and non-interacting (right) categories with RNAPII, H3K4me3 and FoxA1 functional marks. The left y axis is relative tag density for H3K4me3 and RNAPII ChIP-seq data; the right y axis is ChIP-chip intensity for FoxA1 ChIP-chip data. b, Association of proximal and distal interacting (left) and non-interacting (right) ER-αBSs with RNAPII (top) and H3K4me3 (bottom) functional marks.
Next we examined the H3K4me3, RNAPII and FoxA1 marks with respect to ER-αBSs proximal or distal to gene promoters and their involvement in chromatin interactions. Proximal ER-αBSs, whether involved in interactions or not, were highly enriched in H3K4me3, but this was not true of distal ER-αBSs, which was expected because H3K4me3 is a known mark for promoter regions (Fig. 3b and Supplementary Fig. 15b; significance tests are shown in Supplementary Note 5). Proximal ER-αBSs were also highly enriched with RNAPII marks, but the enrichment for both proximal and distal ER-αBSs involved in interactions was significantly higher than that of the proximal and distal ER-αBSs that are not involved in interactions. Intriguingly, although RNAPII showed less enrichment around distal interacting ER-αBSs compared with proximal interacting ER-αBSs, the enrichment was significantly higher than that with distal non-interacting ER-αBSs. Conversely, FoxA1 binding was more enriched around distal ER-αBSs than around proximal ER-αBSs, and most enriched around interacting distal ER-αBSs (Supplementary Fig. 15c), and differences were statistically significant (significance tests are shown in Supplementary Note 5). This indicates that RNAPII and FoxA1, but not H3K4me3, predict interactions at distal ER-αBSs, and suggests that RNAPII and FoxA1 participate in tethering chromatin interactions. Whereas RNAPII is strongly associated with ER-αBSs for transcription activation, FoxA1 is more directly correlated with the regulatory function of ER-α at distal ER-αBSs. At least six interacting ER-αBSs bracket the FOXA1 gene, signifying ER-α-mediated chromatin interactions may regulate FOXA1 (Fig. 2b), further supporting the hypothesis that FoxA1 and ER-α may regulate each other29.
Subsequently, we examined the 689 ER-α-bound chromatin interaction regions with regard to looping structure and gene transcription. We envisage that multiple ER-αBSs may function as ‘anchor’ regions forming chromatin looping structures in three-dimensional space (Fig. 4a). Genes close to interaction anchors are considered to be ‘anchor genes’, and genes in the interaction loop regions and faraway from anchors to be ‘loop genes’. We annotated the interaction regions in relation to UCSC gene database transcripts26 (a gene may have multiple transcripts; here we report transcript numbers, but gene numbers are given in Supplementary Note 6). A gene was considered to be associated with a chromatin interaction region if the TSS of a gene was within 20 kb of the interaction boundaries (Supplementary Fig. 14), a parameter that includes many known and validated ER-α target genes. Most interaction regions (393 out of 689 = 57%) were associated with ‘anchor genes’ (TSS to interaction anchor within 20 kb). Altogether, 1,575 ‘anchor genes’ and 3,767 ‘loop genes’ (TSS more than 20 kb away from interaction anchors) were assigned to interaction regions (Supplementary Tables 3 and 8). Using the same distance parameter (20 kb), we assigned 11,790 genes to 12,126 stand-alone ER-αBSs not involved in interactions (Supplementary Note 6).
a, Distal ER-αBSs interact with proximal sites, forming chromatin loops. Anchor genes (green and blue) are close to interaction anchors with concentrated active transcriptional machinery (red shading). Other genes far from interaction centres (grey) are less active. b, Expression microarray data (oestrogen induction from 0 to 48 h; red denotes activation, and green repression) for interaction anchor genes, loop genes and genes near non-interacting ER-αBSs, with all other UCSC genes26. ‘All genes’ denotes background. c, ChIA-PET interactions data at the FOS/JDP2/BATF loci. Transcription activities are shown by H3K4me3/RNAPII ChIP-Seq and RT–qPCR analysis (bottom panels, the x axis is oestrogen induction time points from 0 to 24 h; the y axis is relative expression (fold increase over ET control)). Error bars show s.e.m.
Within interaction regions with at least one anchor gene there are 1,073 distal ER-αBSs and 387 proximal ER-αBSs (less than 5 kb to TSS), and all distal ER-αBSs (5′ or 3′ to the gene promoter) are looped to anchor genes through connections with proximal ER-αBSs. Many interaction regions include multiple genes, such as the keratin gene cluster (Fig. 1c) and the NR2F2 locus (Fig. 1d), whereas others include only single genes, such as SIAH2 (Fig. 1b). Distal ER-αBSs are stronger than proximal ER-αBSs; this is the inverse of RNAPII marks, which are stronger at gene promoters than at distal regions (Supplementary Fig. 16; examples are shown in Fig. 1 and Supplementary Fig. 17). These observations suggest that direct ER-α binding might be initiated primarily at one or multiple distal sites, which then subsequently recruit other binding sites as anchors to form an interaction complex to ultimately engage the transcriptional machinery at gene promoters.
In addition, we found 296 interaction regions with no associated anchor genes. Although 41 regions contain loop genes, the remaining 255 have no associated UCSC genes assigned to them. Although some interaction regions could be noise or non-functional, some interactions are near gene promoters just outside the 20 kb cutoff, and further sequencing might extend the interaction data to the promoters. The presence of H3K4me3, RNAPII marks and RT–qPCR data at the interaction anchor sites suggests that some interactions could be involved in regulating yet-to-be identified transcripts, such as computationally predicted genes and non-coding RNA species (Supplementary Fig. 18). Alternatively, such interactions could be associated with maintaining chromatin structures or other unknown functions.
To understand whether genes associated with ER-α-bound interactions are regulated by oestrogen, we analysed expression profiles of several interaction-associated genes by RT–qPCR over a time course of oestrogen induction (Supplementary Methods). All anchor genes examined are upregulated by oestrogen induction (Supplementary Fig. 8). We extended our analysis to all interaction-associated genes with the use of whole-genome gene expression microarrays (Fig. 4b). Most ‘anchor genes’ are upregulated (60%), particularly at early time points, in comparison with ‘loop genes’ (48%), indicating that ‘anchor genes’ are significantly associated with gene upregulation (two-tailed P = 1.25 × 10-16; Fig. 4c, Supplementary Note 7, Supplementary Table 9 and Supplementary Fig. 19). In addition, RNAPII marks are associated more with ‘anchor genes’ (39%) than with ‘loop genes’ (26%) (two-tailed P = 10-19). Conversely, genes assigned to ER-αBSs not involved in interactions (on the basis that the gene promoters are within 20 kb of non-interacting ER-αBSs) have very similar expression profiles to that of the background control (all UCSC genes not associated with interactions), indicating that genes associated with non-interacting ER-αBSs are less activated than genes associated with interaction ER-αBSs (significance tests are shown in Supplementary Note 7). Hence, some stand-alone ER-αBSs could be noise, whereas others could involve non-looping mechanisms such as the recruitment of secondary coactivators for downstream functions6.
Within the anchor gene category, many (495 out of 1,575 = 31%) gene entries have 5′ and 3′ ends within interaction boundaries. Such entries, called ‘enclosed anchor genes’, frequently occupy the entirety of short interaction loops, engage multiple anchor sites around or within the gene, tend to have intense RNAPII marks covering the entire gene (examples are shown in Fig. 2b, c and Supplementary Fig. 20) and are preferentially associated with RNAPII marks and gene upregulation as indicated by expression microarrays (Supplementary Note 7 and Supplementary Table 9).
Taken together, our data show an association between chromatin interactions and gene transcriptional activation: enclosed anchor genes are closely correlated with upregulation as measured by gene expression microarray data and RNAPII ChIP-Seq peaks; less closely correlated are non-enclosed anchor genes, loop genes even less so, and genes not associated with interactions are much less so. These results suggest that gene-centric interaction structures may enclose a compartment for concentrating ER-α and transcription-related proteins at target genes.
ER-α-bound interactions may coordinate transcriptional regulation for multiple genes involved in the same functional pathways. At the keratin gene cluster interaction loci (Fig. 1c), enclosed anchor genes such as KRT8 and KRT18 are actively transcribing, as demonstrated by RNAPII and H3K4me3 marks, whereas the loop genes such as KRT72 and KRT75, which are mainly keratins expressed in hair cells that do not have a function in mammary epithelial cells such as MCF-7, are mostly inactive (Supplementary Note 8). Another example is the complex interaction that encompasses the three genes FOS, JDP2 and BATF (Fig. 4c), which encode the dimerization partners of JUN to form the AP-1 transcription factors. AP-1 is important in regulating oestrogen-receptor-dependent transcription by functioning either as a DNA tethering partner or as an ER-α cofactor30. In this complex interaction, FOS and BATF are enclosed anchor genes and are upregulated as shown by RNAPII marks and RT–qPCR, whereas JDP2 is a loop gene and is downregulated as shown by RT–qPCR and decreased RNAPII occupancy. The promoter of JDP2 is marked by H3K4me3, a common feature found in many loop genes (Supplementary Table 9). JDP2 and other loop genes could be ‘poised’ for activation if they were to escape from the interaction loop. Long-range transcriptional regulation by ER-α may therefore be a fine-tuning mechanism that evolved to regulate specific sets of related genes differentially.
To determine functionally whether some ER-α-associated interaction regions are dependent on ER-α, we used short interfering RNA (siRNA) to knock down the level of ER-α protein in MCF-7 cells (Supplementary Methods) and then measured whether the interactions and gene transcription were affected. ER-α-specific siRNA (siER-α) efficiently decreased the amount of ER-α protein and effectively abolished the interactions as demonstrated by a set of 3C assays at the GREB1 locus (Fig. 5). Furthermore, siER-α blocked GREB1 transcription as determined by RT–qPCR. Similar results were also previously shown at the TFF1 site31. Together, these data suggest that at least some of the regulatory long-range chromatin interactions identified by ER-α ChIA-PET data are mediated by ER-α.
a, Genome browser at the GREB1 locus showing data tracks: 1 and 2, H3K4me3 and RNAPII ChIP-Seq; 3 and 4, RNAPII ChIP-qPCR scans using different RNAPII antibodies under oestrogen induction (E2, red) and ethanol control (ET, grey); 5, ER-α (orange) and FoxA1 (green) ChIP-chip7; 6, ChIA-PET density; 7, interaction data. b–d, siRNA knockdown experiments. MCF-7 cells were transfected with siRNA against ER-α (siER-α) or control (siCtrl), and then analysed by western blot analysis with ER-α and calnexin (control) antibodies (b); RT–qPCR on GREB1 expression (c); and 3C assays at GREB1 (d): siER-α knockdown abolishes chromatin interactions and turns off transcription. Error bars show s.e.m. The y axes are relative expression (c) and normalized interaction (d).
Discussion
We demonstrated the ChIA-PET mapping strategy is an unbiased whole-genome approach for the de novo analysis of chromatin interactions, and hence is a major technological advance in our ability to study higher-order organization of chromosomal structures and functions. The ChIA-PET interaction data greatly increase the accuracy of assigning distal TFBSs to target genes, and globally addresses the three-dimensional chromatin interaction mechanism by which distal TFBSs regulate transcription. We postulate the following primary mechanism for ER-α function: ER-α protein dimers are recruited to multiple and primarily distal ER-αBSs, which interact with one another and possibly with other factors such as FoxA1 and RNAPII to form chromatin looping structures around target genes; such topological architectures may partition individual genes into subcompartments of nuclear space such as interaction-anchor-associated genes and interaction-loop-associated genes for differential transcriptional activation or repression. We further speculate that tightly enclosed chromatin interaction centres could help achieve and maintain high local concentration of transcription components for efficient cycling of transcriptional machinery on target gene templates (a summary of results is given in Supplementary Information, and more discussion in Supplementary Note 9).
We expect that this global chromatin interactome map and the ChIA-PET assay will be a valuable starting point for future studies of the three-dimensional architecture of transcription biology in whole-genome contexts.
Methods Summary
MCF-7 cells grown in hormone-depleted medium were treated with 17β-oestradiol (‘oestrogen’, E2) for 45 min before being crosslinked with 1% formaldehyde for 10 min. ChIA-PET libraries were constructed by first performing ChIP with HC-20 antibody (Santa Cruz) or Mab-NRF3A6-050 antibody (Diagenode)10 against ER-α. DNA fragments in ChIP complexes were then ligated to biotinylated half-linkers (linker ligation) containing flanking MmeI restriction sites. The complexes were further ligated under dilute conditions (proximity ligation). PETs were extracted from the ligation products by digestion with MmeI. Released biotinylated PETs were purified by streptavidin-coated magnetic beads, ligated to adaptors, and amplified by PCR. Gel-purified amplicons of PET templates were sequenced by Roche/454 and/or Illumina paired-end sequencing. PET sequences were mapped to the human reference genome (hg18). Binding sites and interactions were identified by using a readout of overlap PETs. To correct for ChIP enrichment bias, we formulated a statistical analysis framework to calculate the probability of the formation of inter-ligation PETs between two regions if ligations between DNA fragments occur by chance. Interactions were further collapsed into complex interactions if they shared interaction anchors. UCSC genes were assigned to interaction regions if they were within 20 kb of interaction regions. To characterize ER-α-bound interactions and associated genes functionally, we conducted gene expression microarray experiments in a time course with and without E2 treatment, and generated genome-wide maps of H3K4me3 (ab8580; Abcam) and RNAPII (serine-5 phosphorylation antibody, ab5131; Abcam) ChIP-Seq data by using Illumina GA single-read sequencing. Interaction-associated genes were annotated with expression microarray data and RNAPII and H3K4me3 ChIP-Seq peaks. Validation experiments included ChIP-qPCR, 3C, ChIP-3C, 4C, FISH and RT–qPCR. For siRNA studies, ER-α ON-TARGETplus SMARTpool siRNA (Dharmacon) was transfected into MCF-7 cells with Lipofectamine 2000 (Invitrogen). Sequences used in experiments are listed in Supplementary Table 10.
References
Fraser, P. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 16, 490–495 (2006)
Collas, P. & Dahl, J. A. Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation. Front. Biosci. 13, 929–943 (2008)
Wei, C. L. et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 (2006)
Wold, B. & Myers, R. M. Sequence census methods for functional genomics. Nature Methods 5, 19–21 (2008)
Massie, C. E. & Mills, I. G. ChIPping away at gene regulation. EMBO Rep. 9, 337–343 (2008)
Carroll, J. S. et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005)
Carroll, J. S. et al. Genome-wide analysis of estrogen receptor binding sites. Nature Genet. 38, 1289–1297 (2006)
Lin, C. Y. et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 3, e87 (2007)
Lupien, M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008)
Welboren, W. J. et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28, 1418–1428 (2009)
West, A. G. & Fraser, P. Remote control of gene transcription. Hum. Mol. Genet. 14 (Review Issue 1) R101–R111 (2005)
Woodcock, C. L. Chromatin architecture. Curr. Opin. Struct. Biol. 16, 213–220 (2006)
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002)
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nature Protocols 2, 1722–1733 (2007)
Horike, S., Cai, S., Miyano, M., Cheng, J. F. & Kohwi-Shigematsu, T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nature Genet. 37, 31–40 (2005)
Cai, S., Lee, C. C. & Kohwi-Shigematsu, T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nature Genet. 38, 1278–1288 (2006)
Zhao, Z. et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genet. 38, 1341–1347 (2006)
Ling, J. Q. et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 (2006)
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature Genet. 38, 1348–1354 (2006)
Wurtele, H. & Chartrand, P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Res. 14, 477–495 (2006)
Dostie, J. et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006)
Tiwari, V. K., Cope, L., McGarvey, K. M., Ohm, J. E. & Baylin, S. B. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 18, 1171–1179 (2008)
Carter, D., Chakalova, L., Osborne, C. S., Dai, Y. F. & Fraser, P. Long-range chromatin regulatory interactions in vivo . Nature Genet. 32, 623–626 (2002)
Osborne, C. S. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature Genet. 36, 1065–1071 (2004)
Simonis, M., Kooren, J. & de Laat, W. An evaluation of 3C-based methods to capture DNA interactions. Nature Methods 4, 895–901 (2007)
Hsu, F. et al. The UCSC Known Genes. Bioinformatics 22, 1036–1046 (2006)
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007)
Phatnani, H. P. & Greenleaf, A. L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 (2006)
Laganiere, J. et al. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl Acad. Sci. USA 102, 11651–11656 (2005)
Kushner, P. J. et al. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74, 311–317 (2000)
Pan, Y. F. et al. Regulation of estrogen receptor-mediated long-range transcription via evolutionarily conserved distal response elements. J. Biol. Chem. 283, 32977–32988 (2008)
Acknowledgements
The authors acknowledge the Genome Technology and Biology Group at the Genome Institute of Singapore for technical support; A. Shahab, C. C. Seng and F. H. Mulawadi for computing support; S. Luo and G. Schroth for Illumina sequencing support; and W. de Laat, B. Ren and X. S. Liu for advice. M.J.F., P.Y.H.H., Y.H., P.Y.T. and Y.K.L. are supported by A*STAR Scholarships. M.J.F. is supported by a L’Oreal-UNESCO For Women In Science National Fellowship. Y.R. and C.L.W. are supported by A*STAR of Singapore and NIH ENCODE grants (R01 HG004456-01, R01HG003521-01 and part of 1U54HG004557-01).
Author Contributions M.J.F. and Y.R. conceptualized the ChIA-PET strategy. M.J.F., E.C. and Y.R. designed the experiments. M.J.F., M.H.L., Y.F.P., J.L., A.H., P.H.M., E.G.Y.C., P.Y.Y.H., W.-J.W., Y.H., Y.L., P.Y.T., P.Y.C., K.D.S.A.W., B.Z., K.S.L., S.C.L., J.S.Y., R.J., K.V.D., J.S.T., Y.K.L., T.H., H.G.S., X.R. and V.C.-R. performed experiments. M.J.F., X.H., Y.B.M., Y.L.O., S.V., H.-S.O., P.N.A., V.B.V., Y.K.L., R.K.M.K., G.B., H.G.S., X.R., V.C.-R., W-K.S., C.-L.W., E.C. and Y.R. analysed experimental data. E.T.L., E.C. and C.-L.W. commented on the manuscript drafts; M.J.F. and Y.R. wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Results, Supplementary Methods, Supplementary Notes, Supplementary Tables 4-7 and 9 (see separate files s2-s6 for Supplementary Tables 1-3, 8 and 10), Supplementary Figures 1-21 with Legends, Supplementary References and a Supplementary Glossary. (PDF 16985 kb)
Supplementary Table 1
This file shows all ERα binding sites found by ChIA-PET and supporting datasets, and associated genes. (XLS 3861 kb)
Supplementary Table 2
This file shows all raw interchromosomal and intrachromosomal inter-ligation PET clusters (duplex interactions) found by ChIA-PET experiments. (XLS 1763 kb)
Supplementary Table 3
This file shows processed interaction regions found by ChIA-PET experiments, and associated genes. (XLS 915 kb)
Supplementary Table 8
This file shows loop, anchor and enclosed anchor genes from ChIA-PET experiments, and their transcriptional characteristics. (XLS 2688 kb)
Supplementary Table 10
This file shows sequence and technical information used in the ChIA-PET and validation experiments. (XLS 43 kb)
Rights and permissions
About this article
Cite this article
Fullwood, M., Liu, M., Pan, Y. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009). https://doi.org/10.1038/nature08497
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08497
This article is cited by
-
Kdm1a safeguards the topological boundaries of PRC2-repressed genes and prevents aging-related euchromatinization in neurons
Nature Communications (2024)
-
Computational methods for analysing multiscale 3D genome organization
Nature Reviews Genetics (2024)
-
Advances in the multimodal analysis of the 3D chromatin structure and gene regulation
Experimental & Molecular Medicine (2024)
-
The potential of epigenetic therapy to target the 3D epigenome in endocrine-resistant breast cancer
Nature Structural & Molecular Biology (2024)
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.