Abstract
Single-cell heterogeneity in cell populations arises from a combination of intrinsic and extrinsic factors1,2,3. This heterogeneity has been measured for gene transcription, phosphorylation, cell morphology and drug perturbations, and used to explain various aspects of cellular physiology4,5,6. In all cases, however, the causes of heterogeneity were not studied. Here we analyse, for the first time, the heterogeneous patterns of related cellular activities, namely virus infection, endocytosis and membrane lipid composition in adherent human cells. We reveal correlations with specific cellular states that are defined by the population context of a cell, and we derive probabilistic models that can explain and predict most cellular heterogeneity of these activities, solely on the basis of each cell’s population context. We find that accounting for population-determined heterogeneity is essential for interpreting differences between the activity levels of cell populations. Finally, we reveal that synergy between two molecular components, focal adhesion kinase and the sphingolipid GM1, enhances the population-determined pattern of simian virus 40 (SV40) infection. Our findings provide an explanation for the origin of heterogeneity patterns of cellular activities in adherent cell populations.
Similar content being viewed by others
Main
Virus infection can be considered the end result of the concerted action of many cellular processes, including endocytosis and the establishment of membrane lipid composition7,8. It is well accepted that these activities display heterogeneity within a population of cells, but the underlying causes of this heterogeneity are not known. We can therefore not predict how such activities behave in a population of cells9.
To study the heterogeneity of these activities, we examined large populations of single monoclonal adherent human cells after three days of growth. These were infected by rotavirus, dengue virus, mouse hepatitis virus (MHV) or SV40. MHV uses clathrin-mediated endocytosis (CME) for infectious entry into the cell10, and SV40 binds to the sphingolipid GM1 for host cell attachment and entry11,12. To analyse CME, we allowed cells to internalize fluorescent transferrin13. To determine the amount of GM1, which is enriched in lipid rafts on the cell surface14, we exposed cells to fluorescent cholera toxin B15.
We observed that the cell-to-cell variation or heterogeneity of these activities displays specific patterns in both cancer- (HeLa and A431) (Fig. 1A) and non-cancer-derived (MCF10A) cells (see below). The patterns could not have been the result of physical constraints on the accessibility of virus particles or fluorescent probes to particular cells, because different types of pattern were observed.
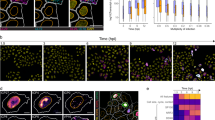
A, GM1 levels on the plasma membrane of A431 cells and dengue virus (DV) infection in HeLa cells are higher in cells at islet edges (upper and lower left panels). MHV infection and CME (upper right and middle panels) are highest in HeLa cells in crowded regions. SV40 (lower right panel) infects HeLa cells that are large and spread out. Scale bars: GM1 and dengue virus, 50 µm; CME, MHV and SV40 respectively 300, 250 and 150 µm. B, Overview of the experimental, computational and statistical methods used in this study (see Methods and Supplementary Methods for details). C, Virus infection (n = 9; grey box-plots) and CME (black lines; average ± single-cell s.d.) plotted against population-context parameters. Model fits are depicted in red. Right axes display the number of cells measured (light-grey bars and lines). D, Regression coefficients between population-context parameters (columns) and cellular activities (rows), colour-coded according to coefficient sign and top-three ranked explanatory power (see legend). ND, not determined. E, Model-predicted virus infection and CME patterns displayed on computer-generated reproductions of original images (left panels). Predicted and measured single-cell activity levels are shown (middle and right panels).
A potential source of single-cell heterogeneity is the deterministic interplay between the phenotypic state of a cell and its activities2,16, mediated by sensors and signal transduction networks. Growing isogenic adherent mammalian cells create cell islets and regions that are sparsely or densely populated, to which cells adapt their size, shape and rate of proliferation17,18 (Supplementary Movie 1), and display several discrete subpopulations19. Together, this implies that patterns of varying cellular states can arise from the adaptation of individual human cells to the particular ‘population context’ in which they reside. The coupling between cell size and timing of meiosis in yeast20, between cell size and the determination of phage infection outcome in Escherichia coli21, and the self-assembly of prokaryotic cells into complex colony patterns22, indicate that such mechanisms also operate in unicellular organisms. We therefore hypothesized that the heterogeneity patterns that we observed are caused by regulatory mechanisms that couple the phenotypic state of a cell to intracellular processes on which virus infection depends, such as endocytosis and the lipid composition of the cell surface.
To reveal the presence of such mechanisms, we used three data-driven modelling approaches on several single-cell measurements obtained by computerized image analysis and supervised machine learning from multiple large populations of cells. (See Fig. 1B for an overview of the methods used, and Supplementary Methods for detailed information.) We first measured for each cell the size of the population to which it belonged, its local cell density, its position on a cell islet edge, its cell size, its mitotic state and its apoptotic state (microenvironment and cell state). By delineating how these parameters interact with each other, we quantified how the characteristics of a cell population are determined. This showed that each parameter, in particular population size, local cell density, position on a cell islet edge, and cell size, represents a population-determined property of an individual cell. The same interactions were observed in cancer cells (Supplementary Fig. 1), normal diploid cells (see below) and primary cells (not shown), despite their different morphologies and growth rates.
For each cell, we next determined whether it was infected, or we quantified its CME activity or cell-surface level of GM1 (cellular activities). We then used the population-determined properties as predictors to model the probability of infection, the activity of CME and the amount of GM1 on the cell surface. In each instance, models could be derived that had good fits (Fig. 1C and Supplementary Fig. 2). The model parameters demonstrated the existence of extensive and specific regulatory mechanisms between population context and virus infection, endocytosis and membrane lipid composition (Fig. 1D). Importantly, the models were able to predict accurately the heterogeneity patterns of cellular activities, solely based on a quantitative assessment of the population context and the state of individual cells (Fig. 1E).
An interesting question that arises is to what extent these population-determined effects contribute to the total variation observed in a population. Strikingly, variance analysis (Supplementary Methods) demonstrated that variation determined by population context is a major component of the total variation observed. For rotavirus, SV40 and dengue virus infection, our measures of the local environment and cellular state account for at least 60%, 74% and 82% of the respective total variation for each activity (Fig. 2a). For CME and GM1 content on the cell surface, variation determined by population context appeared to constitute at least 30% and 21% of the respective total variation (Fig. 2a).
a, Population-determined variance (percentage of total) in the levels of virus infection, CME and GM1 content (Supplementary Methods). Variance of CME in groups of cells in similar population contexts (insert; green) compared with randomized groups of cells (blue). b, Coefficient of variation (s.d./mean) of CME in groups of cells with similar population contexts plotted against local cell density (r2 = -0.57, P < 10-10). Number of cells per group (dot size) and average activity level (colour) are indicated (see legend). c, Significantly different dengue virus infection (P = 2.4 × 10-6) between unperturbed populations of cells with different population properties (n = 9). Model correction abolishes this difference (P = 0.97). d, Significantly different single-cell CME activities (P < 10-10) between unperturbed populations of cells with different population properties. Model correction abolishes this difference (P = 0.75).
To understand better the variation unexplained by population context, we analysed the variance in CME activity in groups of cells with similar population-determined phenotypic properties. This revealed that cells in certain population contexts displayed as much as a 20-fold lower variation than randomly selected cells (inset Fig. 2a). When we analysed this variance relative to the mean CME activity in these groups of cells (coefficient of variation), we observed a threefold lower coefficient of variation in cells with a high local cell density (Fig. 2b). This indicates that these cells have, besides a high CME activity, mechanisms in place that exert a tight control on this activity.
The finding that much of the variation in virus infection, endocytosis and lipid composition of the cell surface is deterministically established by the adaptation of cells to their population context reveals a fundamental problem in our current methods of studying differences in these activities between cell populations. We illustrate this by comparing the fraction of cells infected with dengue virus (Fig. 2c) and the single-cell distributions of CME activity (Fig. 2d) between unperturbed populations of different sizes. The activities display levels that are statistically different (respectively P = 2.4 × 10-6 and P < 10-10) between the different unperturbed populations. However, after normalization of both activities with activities predicted by their respective models, no significant differences were found (P values of respectively 0.97 and 0.75). This is the correct conclusion, as both populations were unperturbed and only differed in their population context. A similar problem exists with supervised and unsupervised clustering methods. Even though such methods correctly identify different phenotypic states (Supplementary Fig. 3), they will not reveal whether these phenotypic states are determined by cellular adaptations to the population context. Therefore, these methods will be unable to distinguish between a change in cellular activity that is a consequence of an altered population context and a change that is a consequence of a direct perturbation.
Besides these fundamental implications, our analysis reveals suggestions for the mechanisms underlying population-determined heterogeneity of virus infection (see ranking in Fig. 1d and Supplementary Fig. 4). The infection efficiency of rotavirus is strongly increased in cells that grow in sparsely populated areas. Indeed, rotavirus uses integrins as its receptor23, which are probably highly expressed on the surface of cells that grow sparsely24. MHV, on the contrary, prefers to infect cells that grow at high local density, similar to where CME is most active. Dengue virus infection occurs almost exclusively in cells located on the edge of cell islets, indicating that it relies on mechanisms highly active in polarized cells25.
The heterogeneity signature of SV40 infection demonstrated a particular preference for large and spread-out cells in both cancer- and non-cancer-derived cell lines (see below). As these cells are predominantly located on the edge of cell islets and in areas that are sparsely populated, they also have higher amounts of GM1 on the cell surface (Fig. 1D), the receptor for SV4011,12. In addition, we previously found that focal adhesion kinase (FAK or PTK2) is required for SV40 infection8, and regulatory links between the microenvironment of cells and FAK24, and between FAK and sphingolipid membrane domains, have been reported26,27.
To test whether FAK and GM1 can be placed in a causal network that determines the heterogeneity pattern of SV40 infection, we applied Bayesian network learning5 (Supplementary Methods) on single-cell measurements of SV40 infection, amount of GM1 on the cell surface, level of Y397-phosphorylated (activated) FAK and population-determined phenotypic properties (Fig. 3a and Supplementary Fig. 5). We chose to use MCF10A cells to point out the relevance of these phenomena also for non-cancer-derived, normal diploid cells. This identified a unique causal network that combines single-cell microenvironmental parameters with molecular components and virus infection (Fig. 3b). We found that cell density is coupled to the regulation of cell size through GM1 cell surface content, and FAK activation. The last two are also determinants of SV40 infection, in addition to as yet unidentified factors that are regulated by cell density. We validated three causal interactions in the Bayesian network. Exogenous addition of GM1 to MCF10A cells increased the levels of phosphorylated FAK and increased SV40 infection. Additionally, knockdown of FAK by RNA interference strongly decreased SV40 infection (Supplementary Fig. 7). Thus, by applying quantitative data-driven modelling on a combination of microenvironmental, cellular and molecular parameters, we reveal mechanistic insights into the establishment of cellular heterogeneity.
a, SV40 infection (red), GM1 content (green) and Y397-phosphorylated FAK (enlargements; yellow) in MCF10A cells. Scale bar, 150 µm. b, Bootstrapped Bayesian network learning (Supplementary Fig. 6 and Supplementary Methods) reveals causal links between population-context parameters, GM1 content, pFAK levels and SV40 infection. Edges are drawn according to the sign of pairwise single-cell correlations. Variation in levels of GM1, pFAK and SV40 infection explained by population context are respectively 30%, 65% and 74%. c, Scatter plot of exogenous GM1-induced increase in pFAK (percentage of non-induced) against local cell density. Linear fit (red) and total number of cells for each measurement (right axis, grey) are shown.
We next wondered why the heterogeneity of SV40 infection is more deterministic (74% explained) than the GM1 levels and FAK activity on which it depends (respectively 30% and 65% explained) (Fig. 3b). Interestingly, when we validated the causal interaction between GM1 and FAK by exogenous addition of GM1, we observed a 2.5-fold stronger activation of FAK in sparse cells compared with dense cells (Fig. 3c). This indicates that GM1 and FAK synergize in sparse cells, leading to the more deterministic downstream activity of SV40 infection. Such synergistic mechanisms can partly cancel out the intrinsic and uncorrelated noise of individual components, increasing the deterministic nature of complex activities at the cellular level2,28.
We have shown that much of the variation in virus infection, endocytosis and membrane lipid composition is determined by the adaptation of cells to their population context, and uncovered basic predictive principles by which non-differentiated cells create complex patterns of activity. Similar mechanisms may determine the heterogeneity of other cellular processes. Perturbation screens, combined with quantitative modelling of single cells in their population context, will further reveal the molecular networks that regulate heterogeneity patterns in cell populations. The principles described here most likely operate in all systems of collective cellular behaviour, from prokaryotic colonies to multicellular organisms.
Methods Summary
All human cell lines were maintained under standard tissue culture conditions. All assays were performed in 96-well plates, and cells were imaged on automated widefield cellWoRx microscopes (Applied Precision) or ImageXpress Micro microscopes (Molecular Devices) with ×10 or ×20 magnification. Infection assays were performed as described (Supplementary Information). CME was measured using Alexa Fluor 488-conjugated transferrin, and GM1 levels were visualized using cholera toxin B conjugated to Alexa Fluor 568 as described8. Phosphorylated-FAK [Y397] was visualized by using standard indirect immunofluorescence protocols. High-content single-cell image analysis was performed using CellProfiler29 and additional image analysis algorithms written specifically for this study (Supplementary Information). In short, cells were identified by object detection on DAPI images, and these regions were typically expanded to cover the cytoplasm. Shape features and features describing the texture and intensities in all channels were extracted for these regions of interest. Support vector machine learning was applied (P. Rämö, R. Sacher, B. Snijder, B. Begemann and L. Pelkmans, submitted) for the classification of diverse cellular phenotypes, including interphase, mitotic and apoptotic cellular states, as well as infectious phenotypes and technical artefacts. Globally, these data were further analysed using several statistical methods. First, graphical Gaussian modelling quantified how a population shapes the property distributions of individual cells and how individual properties influence each other, which was experimentally validated by live cell imaging. Second, probit regression modelling revealed how virus infection depends on population-determined properties of individual cells, as did weighted linear regression for GM1 content and CME. Finally, bootstrapped Bayesian network learning was applied to identify causal interactions between properties of the microenvironment, FAK-phosphorylation, GM1 content and SV40 infection in MCF10A cells. Selected inferred causal links were experimentally validated by using GM1 addback and short interfering RNA (siRNA)-mediated silencing of FAK as described8. A detailed description of the methods performed in this study is provided in the Supplementary Information.
References
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002)
Maheshri, N. & O’Shea, E. K. Living with noisy genes: how cells function reliably with inherent variability in gene expression. Annu. Rev. Biophys. Biomol. Struct. 36, 413–434 (2007)
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008)
Sigal, A. et al. Variability and memory of protein levels in human cells. Nature 444, 643–646 (2006)
Sachs, K., Perez, O., Pe’er, D., Lauffenburger, D. A. & Nolan, G. P. Causal protein-signaling networks derived from multiparameter single-cell data. Science 308, 523–529 (2005)
Keren, K. et al. Mechanism of shape determination in motile cells. Nature 453, 475–480 (2008)
Liberali, P., Ramo, P. & Pelkmans, L. Protein kinases: starting a molecular systems view of endocytosis. Annu. Rev. Cell Dev. Biol. 24, 501–523 (2008)
Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78–86 (2005)
Wilkinson, D. J. Stochastic modelling for quantitative description of heterogeneous biological systems. Nature Rev. Genet. 10, 122–133 (2009)
Eifart, P. et al. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 81, 10758–10768 (2007)
Neu, U., Woellner, K., Gauglitz, G. & Stehle, T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl Acad. Sci. USA 105, 5219–5224 (2008)
Pelkmans, L. Secrets of caveolae- and lipid raft-mediated endocytosis revealed by mammalian viruses. Biochim. Biophys. Acta 1746, 295–304 (2005)
Conner, S. D. & Schmid, S. L. Regulated portals of entry into the cell. Nature 422, 37–44 (2003)
Mayor, S. & Pagano, R. E. Pathways of clathrin-independent endocytosis. Nature Rev. Mol. Cell Biol. 8, 603–612 (2007)
Holmgren, J., Lonnroth, I. & Svennerholm, L. Tissue receptor for cholera exotoxin: postulated structure from studies with GM1 ganglioside and related glycolipids. Infect. Immun. 8, 208–214 (1973)
Sacher, R., Stergiou, L. & Pelkmans, L. Lessons from genetics: interpreting complex phenotypes in RNAi screens. Curr. Opin. Cell Biol. 20, 483–489 (2008)
Eagle, H. & Levine, E. M. Growth regulatory effects of cellular interaction. Nature 213, 1102–1106 (1967)
Castor, L. N. Flattening, movement and control of division of epithelial-like cells. J. Cell. Physiol. 75, 57–64 (1970)
Slack, M. D., Martinez, E. D., Wu, L. F. & Altschuler, S. J. Characterizing heterogeneous cellular responses to perturbations. Proc. Natl Acad. Sci. USA 105, 19306–19311 (2008)
Nachman, I., Regev, A. & Ramanathan, S. Dissecting timing variability in yeast meiosis. Cell 131, 544–556 (2007)
St-Pierre, F. & Endy, D. Determination of cell fate selection during phage lambda infection. Proc. Natl Acad. Sci. USA 105, 20705–20710 (2008)
Ben-Jacob, E., Cohen, I. & Gutnick, D. L. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu. Rev. Microbiol. 52, 779–806 (1998)
Lopez, S. & Arias, C. F. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 12, 271–278 (2004)
Geiger, B., Spatz, J. P. & Bershadsky, A. D. Environmental sensing through focal adhesions. Nature Rev. Mol. Cell Biol. 10, 21–33 (2009)
Neumann, A. K., Thompson, N. L. & Jacobson, K. Distribution and lateral mobility of DC-SIGN on immature dendritic cells–implications for pathogen uptake. J. Cell Sci. 121, 634–643 (2008)
Iwabuchi, K. et al. Reconstitution of membranes simulating “glycosignaling domain” and their susceptibility to Lyso-GM3. J. Biol. Chem. 275, 15174–15181 (2000)
Palazzo, A. F., Eng, C. H., Schlaepfer, D. D., Marcantonio, E. E. & Gundersen, G. G. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303, 836–839 (2004)
Newman, J. R. & Weissman, J. S. Systems biology: many things from one. Nature 444, 561–562 (2006)
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006)
Acknowledgements
We acknowledge H. Verheije and L. Burleigh for providing images of MHV and dengue virus infection, G. Jurisic for providing primary cells and help with experiments, and all members of the laboratory for comments on the manuscript. P.R. is supported by the European Molecular Biology Organisation and the Human Frontiers Science Program, E.-M.D. by Oncosuisse and P.L. by the Federation of European Biochemical Societies. L.P. is supported by the ETH Zürich, SystemsX.ch, the Swiss National Science Foundation and the European Union.
Author Contributions L.P. supervised and conceived the project. R.S., B.S., E.-M.D. and P.L. performed experiments, B.S. and P.R. developed computational image analysis methods, B.S. performed all computational image analysis, B.S. and P.R. conceived the statistical analysis methods, B.S. performed all statistical analysis, L.P. and B.S. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10 with Legends, Supplementary Movie 1 Legend, Supplementary Methods, Supplementary Data, Supplementary Table 1 and Supplementary References. (PDF 6784 kb)
Supplementary Movie 1
This movie file shows that population properties are determined during growth of adherent human cells - see file s1 for full Legend. (MOV 9511 kb)
Rights and permissions
About this article
Cite this article
Snijder, B., Sacher, R., Rämö, P. et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520–523 (2009). https://doi.org/10.1038/nature08282
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08282
This article is cited by
-
Cellular state landscape and herpes simplex virus type 1 infection progression are connected
Nature Communications (2023)
-
Research on GGT-responsive drug carrier with active transport effect
Nano Research (2023)
-
Deciphering variations in the endocytic uptake of a cell-penetrating peptide: the crucial role of cell culture protocols
Cytotechnology (2023)
-
Relating individual cell division events to single-cell ERK and Akt activity time courses
Scientific Reports (2022)
-
In plaque-mass spectrometry imaging of a bloom-forming alga during viral infection reveals a metabolic shift towards odd-chain fatty acid lipids
Nature Microbiology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.