Abstract
Molecular self-assembly offers a ‘bottom-up’ route to fabrication with subnanometre precision of complex structures from simple components1. DNA has proved to be a versatile building block2,3,4,5 for programmable construction of such objects, including two-dimensional crystals6, nanotubes7,8,9,10,11, and three-dimensional wireframe nanopolyhedra12,13,14,15,16,17. Templated self-assembly of DNA18 into custom two-dimensional shapes on the megadalton scale has been demonstrated previously with a multiple-kilobase ‘scaffold strand’ that is folded into a flat array of antiparallel helices by interactions with hundreds of oligonucleotide ‘staple strands’19,20. Here we extend this method to building custom three-dimensional shapes formed as pleated layers of helices constrained to a honeycomb lattice. We demonstrate the design and assembly of nanostructures approximating six shapes—monolith, square nut, railed bridge, genie bottle, stacked cross, slotted cross—with precisely controlled dimensions ranging from 10 to 100 nm. We also show hierarchical assembly of structures such as homomultimeric linear tracks and heterotrimeric wireframe icosahedra. Proper assembly requires week-long folding times and calibrated monovalent and divalent cation concentrations. We anticipate that our strategy for self-assembling custom three-dimensional shapes will provide a general route to the manufacture of sophisticated devices bearing features on the nanometre scale.
Similar content being viewed by others
Main
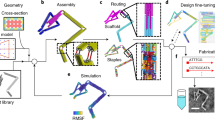
The assembly of a target three-dimensional shape using the honeycomb-pleat-based strategy described here can be conceptualized as laying down the scaffold strand into an array of antiparallel helices (Fig. 1a) where helix m + 1 has a preferred attachment angle to helix m of ±120° degrees with respect to the attachment of helix m - 1 to helix m (Fig. 1b, c). This angle is determined by the relative register along the helical axes of the Holliday-junction crossovers that connect helix m + 1 to helix m versus those that connect helix m - 1 to helix m. Branching flaps are allowed as well (Supplementary Note S1).
a, Double helices comprised of scaffold (grey) and staple strands (orange, white, blue) run parallel to the z-axis to form an unrolled two-dimensional schematic of the target shape. Phosphate linkages form crossovers between adjacent helices, with staple crossovers bridging different layers shown as semicircular arcs. b, Cylinder model of a half-rolled conceptual intermediate. Cylinders represent double helices, with loops of unpaired scaffold strand linking the ends of adjacent helices. c, Cylinder model of folded target shape. The honeycomb arrangement of parallel helices is shown in cross-sectional slices (i–iii) parallel to the x–y plane, spaced apart at seven base-pair intervals that repeat every 21 base pairs. All potential staple crossovers are shown for each cross-section. d, Atomistic DNA model of shape from c.
The design procedure is analogous to sculpture from a porous crystalline block. Here the block is a honeycomb lattice of antiparallel scaffold helices (Fig. 1d). Complementary staple strands wind in an antiparallel direction around the scaffold strands to assemble B-form double helices that are assigned initial geometrical parameters (that can later be adjusted to account for interhelical repulsion) of 2.0 nm diameter, 0.34 nm per base-pair rise, and 34.3° per base-pair mean twist (or 21 base pairs every two turns). Crossovers between adjacent staple helices are restricted to intersections between the block and every third layer of a stack of planes orthogonal to the helical axes, spaced apart at intervals of seven base pairs or two-thirds of a turn (Fig. 1c). Crossovers between adjacent scaffold helices are permitted at positions displaced upstream or downstream of the corresponding staple-crossover points by five base pairs or a half-turn.
The first steps in the design process are carving away duplex segments from the block to define the target shape, and then introducing scaffold crossovers at a subset of allowed positions so as to create a singular scaffold path that visits all remaining duplex segments. Next, staple crossovers are added at all permitted positions on the shape that are not five base pairs away from a scaffold crossover; this exception maintains the local crossover density along any helix–helix interface at roughly one per 21 base pairs. Nicks are introduced into staple helices to define staple strands whose lengths are between 18 and 49 bases inclusive, with a mean between 30 and 42 bases. Sometimes staple crossovers are removed at the edges of the shapes to allow adjustment of staple lengths to preferred values. Unpaired scaffold bases are often introduced at the ends of helices to minimize undesired multimerization, or else to accommodate later addition of connecting staple strands that mediate desired multimerization. The final step is to thread the actual scaffold sequence on the target scaffold path to determine the Watson–Crick-complementary sequences of the staple strands.
Design steps and assignment of staple sequences for the shapes presented here were aided by manual rendering of strand diagrams in Adobe Illustrator and by writing ad hoc computer programs to produce staple sequences corresponding to those diagrams. This process was very time-consuming and error-prone even for trained DNA nanotechnologists. More recently, we have developed caDNAno, a graphical-interface-based computer-aided-design environment for assisting in honeycomb-pleated-origami design21, and have ported all the objects described in this article into this framework (Supplementary Note S2). With caDNAno, an individual with no prior knowledge of programming or DNA structure can complete a short tutorial and then be capable of generating sequences within a day for building a new shape comparable in complexity to the examples demonstrated here.
As with flat DNA origami19, assembly of three-dimensional, honeycomb-pleated DNA origami proceeds in a one-pot reaction, after rapid heating followed by slow cooling, between a scaffold strand and the hundreds of oligonucleotide staple strands that direct its folding into the target shape. Successful folding was observed for a panel of five structural targets (detailed schematics in Supplementary Note S2) each produced by mixing 10 nM scaffold strands derived from the single-stranded genome of the M13 bacteriophage (preparation described in Supplementary Note S1), 50 nM of every oligonucleotide staple strand, purified by reverse-phase cartridge (Bioneer Inc.), buffer and salts including 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C), 16 mM MgCl2, and subjecting the mixture to a thermal-annealing ramp that cooled from 80 °C to 60 °C over the course of 80 min and then cooled from 60 °C to 24 °C over the course of 173 h. Objects were electrophoresed on a 2% agarose gel containing 45 mM Tris borate + 1 mM EDTA (pH 8.3 at 20 °C), and 11 mM MgCl2 at 70 V for four hours cooled by an ice-water bath, monomer bands were excised, DNA was recovered by physical extraction from the excised band, and the objects were imaged using transmission electron microscopy after negative-staining by uranyl formate. The fraction of scaffold strands that were incorporated into monomeric species after folding varied from 7% to 44% for these targets as estimated by ethidium-bromide fluorescence intensity. Gel-purified particles were generally observed to be monodisperse with a homogenous shape (Fig. 2f); defect analysis for a series of related objects can be found elsewhere21.
The first and second rows show perspective and projection views of cylinder models, with each cylinder representing a DNA double helix. a, Monolith. b, Square nut. c, Railed bridge. d, Slotted cross. e, Stacked cross. Rows three to seven show transmission electron microscope (TEM) micrographs of typical particles. For imaging, samples were adsorbed (5 min) onto glow-discharged grids pre-treated with 0.5 M MgCl2, stained with 2% uranyl formate, 25 mM NaOH (1 min), and visualized with an FEI Tecnai T12 BioTWIN at 120 kV. f, Top, field of homogeneous and monodisperse stacked-cross particles. Bottom, expanded view of boxed area from above. g, Left, typical monolith particle. Right, integrated-intensity profile (red) of line orthogonal to the longitudinal axis of typical monolith particle, with expected profile (grey) assuming a simple homogeneous cylinder model. h, Left, gaussian-fitted mean peak positions (circles) in such integrated-line profiles for twenty different monolith particles as a function of peak index. The observed mean peak-to-peak distance was 3.65 nm (±0.2 nm s.d., ±0.01 nm standard error of the mean, s.e.m.). This peak-to-peak distance should correspond to 1.5 times the effective diameter d of individual double helices in the monolith structure, hence d = 2.4 nm. Solid line is a linear fit with a slope of 3.65 nm from peak to peak, corroborating equidistant arrangement of helices across the entire particle width. Error bars (red) indicate mean width of the peaks. Slightly higher variations in peak width at the edges of the particles are most likely due to frayed edges (compare with particles in a and g). i, Analysis as in g repeated for the square-nut shape. j, Histogram of gaussian-fitted peak-to-peak distances as found for the square-nut particles, with the mean value at 3.18 nm (±0.2 nm s.d., ±0.01 nm s.e.m.), indicating an effective diameter of 2.1 nm per individual double helix. a.u., arbitrary units. Scale bars: a–e, 20 nm; f, 1 µm (top), 100 nm (bottom).
The five objects displayed in Fig. 2 demonstrate the generality of this honeycomb-pleated origami approach in approximating various three-dimensional shapes. Figure 2a shows a structure resembling a monolith, assembled in the form of a honeycomb-pleated block as in Fig. 1, except with ten layers instead of three. Particles display the predicted pattern of holes and stripes consistent with a honeycomb lattice of cylinders. Figure 2b shows a square nut, the cross-section of which is a block of the honeycomb lattice with an internal pore shaped like a six-pointed star. Figure 2c shows a structure that resembles a bridge with hand rails. This shape demonstrates that different cross-section patterns can be implemented along the helical axis. Figure 2d shows a slotted cross, a structure composed of two honeycomb-lattice-based domains that sit at 90° to one another. One domain is H-shaped, the other is O-shaped. The centre of the H-domain passes through the slot of the O-domain, and the two domains are connected by a pair of Holliday-junction crossovers derived from the scaffold strand. The 90° angle between domains is enforced by steric collisions between the ends of helices on the H-domain and the sides of helices on the O-domain. The fifth particle image for the slotted cross in Fig. 2d shows a defective particle, where the slot in the O-domain can be seen clearly. Figure 2e shows a stacked cross, where again two domains sit at 90° to one another. One domain is C-shaped, the other domain resembles a pod with a cavity. The pod domain consists of four sub-modules that are each connected to the C-shaped domain by a Holliday-junction crossover derived from the scaffold strand. Upon folding, the sub-modules connect to each other by staple linkages, enforcing a rotation to yield the complete pod domain oriented 90° to the C-module.
For the monolith, an effective diameter of 2.4 nm (±0.1 nm standard deviation, s.d.) per individual double helix was observed (Fig. 2g, h), while for the square nut an effective diameter of 2.1 nm (±0.1 nm s.d.) per individual double helix was observed (Fig. 2i, j). Assuming an unhydrated helical diameter of 2.0 nm (although the hydrodynamic helical diameter has been estimated22 as 2.2–2.6 nm), this observation suggests the presence of inter-helical gaps produced by electrostatic repulsion8 of the order of 0.1–0.4 nm, significantly less than the 1.0 nm gap size estimated for Rothemund flat origami. This discrepancy is probably related to the roughly twofold higher density of crossovers present in the honeycomb-pleated origami. Differences in effective helix diameter between architectures may originate in part from staining artefacts (for example, cavities where large amounts of positively charged stain accumulate, or flattening).
Three key determinants for folding of honeycomb-pleated origami were investigated: duration of thermal ramp, divalent-cation concentration, and monovalent-cation concentration. Folding with short thermal ramps (Fig. 3b, lefthand lanes), low concentrations of MgCl2 (Fig. 3d, lefthand lanes), or high concentrations of NaCl (Fig. 3f, lefthand lanes) yielded a slowly migrating species upon agarose-gel electrophoresis and grossly misshapen objects as observed by transmission electron microscopy (for example, see Fig. 3c). In contrast, week-long thermal annealing at higher concentrations of MgCl2 combined with low concentrations of NaCl yielded a fast-migrating species upon agarose-gel electrophoresis and well-folded particles as observed by electron microscopy (Fig. 3e), along with lower mobility bands corresponding to multimerized and aggregated objects. The apparent trend was that increasing agarose-gel mobility correlated with improvement of quality of folding as observed by transmission electron microscopy, suggesting that correctly folded structures tend to be more compact than misfolded versions.
a, Cylinder models of shapes: monolith, stacked cross, railed bridge, and two versions of genie bottle, with corresponding scaffold sequences. Labels indicate the source of scaffold used for folding the object (for example, p7560 is an M13-based vector of length 7,560 bases). b, Shapes were folded in 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C) and 16 mM MgCl2 and analysed by gel electrophoresis (2% agarose, 45 mM Tris borate + 1 mM EDTA (pH 8.3 at 20 °C), 11 mM MgCl2) using different thermal-annealing ramps. For the 1.2 h ramp, the temperature was lowered from 95 °C to 20 °C at a rate of 1.6 min °C-1. For the 3 h, 6 h, 12 h, 18 h, 37 h, 74 h and 173 h ramps, the temperature was lowered from 80 °C to 60 °C at 4 min °C-1, and then from 60 °C to 24 °C at rates of 5, 10, 20, 30, 60, 120 or 280 min °C-1, respectively). c–e, TEM and gel analysis of influence of MgCl2 concentration on folding quality. c, The fastest-migrating bands in the 4 mM MgCl2 lanes were purified and imaged, revealing gross folding defects. d, Shapes were folded with a 173 h ramp in 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C) and MgCl2 concentrations varying from 0 to 30 mM. e, As in c, leading bands were purified from the 16 mM MgCl2 lanes and found to exhibit higher-quality folding when analysed by TEM. f, Excess NaCl inhibits proper folding. Shapes were folded with 173 h ramp in 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C), 16 mM MgCl2, and varying NaCl concentrations.
Divalent cations thus appear to accelerate the rate of proper folding and increase the amount of undesired aggregation whereas monovalent cations appear to decelerate the rate of proper folding and decrease the amount of undesired aggregation. Many of the structures require week-long thermal ramps for proper folding, even under idealized divalent- and monovalent-cation concentrations. Divalent cations may accelerate target folding by specific stabilization of Holliday-junction crossovers23 and by nonspecific stabilization of compact DNA24 folding intermediates, although they may also stabilize nontarget aggregates by a similar mechanism. Monovalent-cation binding might compete with divalent-cation binding, and thereby antagonize both target compaction and nontarget aggregation, analogous to how such binding inhibits multivalent-cation-induced DNA condensation25. Folding of simpler DNA-origami structures such as the six-helix-bundle nanotube is much more robust to variations in annealing conditions (Supplementary Note S1); the Rothemund flat origami and these simpler nanotube structures could be folded with 72 min ramps. Presumably, multilayered structures must traverse more difficult kinetic traps, perhaps owing in part to the larger density of crossovers, in part to issues of local folding and unfolding in the confined space between two or more layers of DNA helices, and in part to the difficulties in reaching a high density of DNA in the final folded object, similar to that found in high-pressure virus capsids26.
One of the target shapes presented in Fig. 3 — the genie bottle (strand diagram in Supplementary Note S2) — was folded with two different scaffold sequences. Its full size takes up only 4,500 base pairs. One scaffold sequence used for folding was a modified M13 genome with a length of 7,308 bases, where 2,800 bases of the scaffold strand were left unpaired and dangling from the neck of the bottle (reminiscent of wisps of smoke in TEM images), while the other scaffold sequence used was the 4,733-base forward strand of an expression vector encoding the enhanced green fluorescent protein (pEGFP-N1, Clontech). Folding of the same shape under identical conditions gave superior yield with the M13-based scaffold sequence. Some folding success could be achieved with the pEGFP-N1 scaffold sequence when much higher scaffold and staple concentrations were used. One striking difference between the two scaffold sequences is that the M13 base composition is 43% cytosines and guanines whereas the pEGFP-N1 base composition is 53% cytosines and guanines. Higher levels of GC base pairs might lead to a greater incidence of mispairing during folding and a slower rate of unpairing in misfolded intermediates, which could explain why folding was more difficult with the pEGFP-N1 scaffold sequence. On the other hand, local sequence diversity is potentially greatest at 50% GC content, and so a scaffold sequence with GC content that is very low might not be well-suited for DNA origami. Systematic studies will be required in the future to determine the optimal base composition.
Hierarchical assembly of DNA-origami nanostructures can be achieved by programming staple strands to bridge separate scaffold strands. Figure 4a shows the stacked cross programmed to polymerize along the long axes of the DNA helices of the pod domain. The scaffold loops on the ends of the object were programmed with a length such that they form properly spaced scaffold crossovers in the presence of bridging staple strands that link the two ends of the objects. This induces head-to-tail polymerization. Shown are filaments that adsorbed on the grid in two different orientations to illustrate the periodic presentation of the C-shaped domain perpendicular to the filament axis at a periodicity of 41 nm (±3 nm s.d. over a 33mer), corresponding to a length per base pair of 0.33 nm (±0.02 nm s.d.).
a, Left panel, Cylinder model of stacked-cross monomer (Fig. 2e), with dotted line indicating direction of assembly. Right panels, typical TEM micrographs showing stacked-cross polymers. Purified stacked-cross samples were mixed with a fivefold molar excess of connector staple strands in the presence of 5 mM Tris + 1 mM EDTA (pH 7.9 at 20 °C), 16 mM MgCl2 at 30 °C for 24 h. Monomers were folded in separate chambers, purified, and mixed with connector staple strands designed to bridge separate monomers. b, Cylinder model (left) and transmission electron micrograph (right) of a double-triangle shape comprised of 20 six-helix bundle half-struts. c, Heterotrimerization of the icosahedra was done with a 1:1:1 mixture of the three unpurified monomers at 50 °C for 24 h. d, Orthographic projection models and TEM data of four icosahedron particles. Scale bars in a, b and d: 100 nm.
Figure 4b shows a wireframe DNA-origami nanostructure whose struts are six-helix-bundle nanotubes (strand diagrams in Supplementary Note S2). A single scaffold strand is folded into a branched tree that links two pairs of half-struts internally to produce a double triangle (Fig. 4b). This operation is repeated twice more with two completely different sets of staple strands, based on cyclic permutation of the same 8,100-base scaffold sequence through the architecture of the double-triangle monomer. This produces three chemically distinct double-triangle monomers that vary according to the sequences displayed at various positions. Every double triangle displays ten terminal branches presenting scaffold and staple sequences that are programmed to pair specifically with five terminal branches each on the two other double triangles (Fig. 4c). When the three species are mixed together, heterotrimers in the shape of a wireframe icosahedron with a diameter of about 100 nm are formed (Fig. 4d, and gel in Supplementary Note S1). The majority of particles visualized by transmission electron microscopy have missing struts, owing either to incomplete folding or to particle flattening and collapse, commonly seen for spherical or cylindrical particles prepared by negative-stain protocols27.
Previously, scaffolded DNA origami was employed to create flat structures containing dozens of helices and nanotubes containing six helices9,28,29. The present work generalizes this method into three dimensions by folding helices on a honeycomb lattice. Using caDNAno21, staple sequences for folding newly conceived objects can be generated quickly. Design, acquisition of commercially synthesized staple strands, thermal folding, and initial transmission-electron-microscopic imaging can all be completed in as little as two weeks.
Improvements in the rate and yield of folding will be critical for enabling the robust assembly of larger and more complicated DNA nanostructures. Potential steps in this direction include enzymatic synthesis for higher-quality staple strands, artificial scaffold sequences that are more amenable to robust folding, folding with formamide dilution instead of thermal ramps to reduce thermal damage to the DNA29, and hierarchical assembly with monomer architectures that have been identified as being particularly well-behaved.
Three-dimensional origami structures should expand the range of possible applications with an increased range of spatial positioning that is not accessible by flat structures, including those requiring encapsulation or space-filling functionalities. For example, many natural biosynthetic machines, such as polymerases, ribosomes, chaperones, and modular synthases, use three-dimensional scaffolding to control assembly of complex products. Similar capabilities for synthetic machines are thus more accessible with this convenient, generalizable facility to fabricate custom-shaped three-dimensional structures from DNA.
References
Whitesides, G. M., Mathias, J. P. & Seto, C. T. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science 254, 1312–1319 (1991)
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982)
Fu, T. J. & Seeman, N. C. DNA double-crossover molecules. Biochemistry 32, 3211–3220 (1993)
Li, X. J., Yang, X. P., Qi, J. & Seeman, N. C. Antiparallel DNA double crossover molecules as components for nanoconstruction. J. Am. Chem. Soc. 118, 6131–6140 (1996)
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003)
Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. Design and self-assembly of two-dimensional DNA crystals. Nature 394, 539–544 (1998)
Yan, H., Park, S. H., Finkelstein, H., Reif, J. H. & LaBean, T. H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003)
Rothemund, P. W. et al. Design and characterization of programmable DNA nanotubes. J. Am. Chem. Soc. 126, 16344–16352 (2004)
Mathieu, F. et al. Six-helix bundles designed from DNA. Nano Lett. 5, 661–665 (2005)
Liu, D., Park, S. H., Reif, J. H. & LaBean, T. H. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl Acad. Sci. USA 101, 717–722 (2004)
Yin, P. et al. Programming DNA tube circumferences. Science 321, 824–826 (2008)
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005)
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991)
Zhang, Y. & Seeman, N. C. The construction of a DNA truncated octahedron. J. Am. Chem. Soc. 116, 1661–1669 (1994)
He, Y. et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452, 198–201 (2008)
Zhang, C. et al. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc. Natl Acad. Sci. USA 105, 10665–10669 (2008)
Shih, W. M., Quispe, J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004)
Whitesides, G. M. & Grzybowski, B. Self-assembly at all scales. Science 295, 2418–2421 (2002)
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006)
Yan, H., LaBean, T. H., Feng, L. & Reif, J. H. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl Acad. Sci. USA 100, 8103–8108 (2003)
Douglas, S. M. et al. Rapid prototyping of three-dimensional DNA-origami shapes with caDNAno. Nucleic Acids Res (in the press)
Mandelkern, M., Elias, J. G., Eden, D. & Crothers, D. M. The dimensions of DNA in solution. J. Mol. Biol. 152, 153–161 (1981)
Diekmann, S. & Lilley, D. M. J. The anomalous gel migration of a stable cruciform: temperature and salt dependence, and some comparisons with curved DNA. Nucleic Acids Res. 15, 5765–5774 (1987)
Budker, V., Trubetskoy, V. & Wolff, J. A. Condensation of nonstoichiometric DNA/polycation complexes by divalent cations. Biopolymers 83, 646–657 (2006)
Hibino, K. et al. Na+ more strongly inhibits DNA compaction by spermidin(3+) than K+. Chem. Phys. Lett. 426, 405–409 (2006)
Garcia, H. G. et al. Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity. Biopolymers 85, 115–130 (2007)
Harris, J. R., Gerber, M., Gebauer, W., Wernicke, W. & Markl, J. Negative stains containing trehalose: application to tubular and filamentous structures. Microsc. Microanal. 2, 43–52 (1996)
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2007)
Jungmann, R., Liedl, T., Sobey, T. L., Shih, W. & Simmel, F. C. Isothermal assembly of DNA origami structures using denaturing agents. J. Am. Chem. Soc. 130, 10062–10063 (2008)
Acknowledgements
We thank X. Su for assistance in cloning M13-based scaffold sequences and G. Hess for pilot studies on the railed-bridge design. This work was supported by a Claudia Adams Barr Program Investigator grant, a Wyss Institute for Biologically Inspired Engineering at Harvard grant, and an NIH New Investigator grant (1DP2OD004641-01) to W.M.S., a Humboldt Fellowship to H.D., Deutscher Akademischer Austauschdienst (DAAD) Fellowship to T.L., and Swedish Science Council (Vetenskapsrådet) Fellowship to B.H.
Author Contributions S.M.D. designed the monolith and square nut, and provided caDNAno software support; H.D. designed the stacked cross; T.L. designed the railed bridge; B.H. designed the slotted cross; F.G. designed the genie bottle; W.M.S. designed the icosahedron; S.M.D. and W.M.S. developed the honeycomb-pleated-origami design rules; H.D., S.M.D., T.L., B.H. and W.M.S. optimized the folding and imaging conditions. All authors collected and analysed data and contributed to preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
[Competing interests: W.M.S. is listed as inventor on a patent filed by Dan-Farber cancer institutes, entitled ‘Wireframe Nanostructures’ for the wireframe icosahedron described in Fig. 4, in April 2007.]
Additional information
The authors declare competing financial interests: details accompany the paper on www.nature.com/nature.
Supplementary information
Supplementary Information
This file contains Figures s1-s12 with Legends, Supplementary Materials and Methods and Supplementary Data (PDF 13130 kb)
Rights and permissions
About this article
Cite this article
Douglas, S., Dietz, H., Liedl, T. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). https://doi.org/10.1038/nature08016
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08016
This article is cited by
-
Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force
Nature Communications (2024)
-
Harnessing a paper-folding mechanism for reconfigurable DNA origami
Nature (2023)
-
Digital nanoreactors to control absolute stoichiometry and spatiotemporal behavior of DNA receptors within lipid bilayers
Nature Communications (2023)
-
Site-directed placement of three-dimensional DNA origami
Nature Nanotechnology (2023)
-
Gene-encoding DNA origami for mammalian cell expression
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.