Abstract
The diagnosis of a uterine smooth muscle lesion is, in the majority of cases, straightforward. However, in a small number of cases, the morphological criteria used in such lesions cannot differentiate with certainty a benign from a malignant lesion and a diagnosis of smooth muscle tumor with uncertain malignant potential (STUMP) is made. Uterine leiomyosarcomas are often easy to diagnose but it is difficult or even impossible to identify a prognostic factor at the moment of the diagnosis with the exception of the stage. We hypothesize, for uterine smooth muscle lesions, that there is a gradient of genomic complexity that correlates to outcome. We first tested this hypothesis on STUMP lesions in a previous study and demonstrated that this 'gray category' could be split according to genomic index into two groups. A benign group, with a low to moderate alteration rate without recurrence and a malignant group, with a highly rearranged profile akin to uterine leiomyosarcomas. Here, we analyzed a large series of 77 uterine smooth muscle lesions (from 76 patients) morphologically classified as 19 leiomyomas, 14 STUMP and 44 leiomyosarcomas with clinicopathological and genomic correlations. We confirmed that genomic index with a cut-off=10 is a predictor of recurrence (P<0.0001) and with a cut-off=35 is a marker for poor overall survival (P=0.035). For the tumors confined to the uterus, stage as a prognostic factor was not useful in survival prediction. At stage I, among the tumors reclassified as molecular leiomyosarcomas (ie, genomic index ≥10), the poor prognostic markers were: 5p gain (overall survival P=0.0008), genomic index at cut-off=35 (overall survival P=0.0193), 13p loss including RB1 (overall survival P=0.0096) and 17p gain including MYOCD gain (overall survival P=0.0425). Based on these findings (and the feasibility of genomic profiling by array-comparative genomic hybridization), genomic index, 5p and 17p gains prognostic value could be evaluated in future prospective chemotherapy trials.
Similar content being viewed by others
Main
Uterine sarcomas are rare neoplasms accounting for 4% of all uterine tumors with an estimated incidence of 0.86 per 100 000 women.1 Uterine smooth muscle lesions include very common benign tumors such as leiomyomas and malignant as leiomyosarcomas.2 The overall incidence of leiomyomas is 70–80% by 50 years of age.3 Leiomyomas have a benign behavior despite the benign metastasizing leiomyoma and leiomyomatoses, whereas leiomyosarcomas eventually lead to death through recurrences in up to 75% of cases.4, 5
The diagnosis of uterine smooth muscle lesions, based on the Stanford criteria (three morphological features: presence of cytologic atypia, mitotic count and tumor cell necrosis),6 is straightforward in the majority of the cases. However, sometimes morphology is confusing and introduces a degree of subjectivity in the interpretation of these criteria. External factors such as prior treatments (hormonal therapies) or a non-optimal fixation of the sample may pose diagnostic challenges. Hence, such lesions are usually classified as smooth muscle tumors of uncertain malignant potential (STUMP).2 This classification could result in a risk of under or over diagnosis with clinical consequences for the treatment that could impact the patient.
The FIGO stage is still the most important uterine leiomyosarcomas prognostic factor and forms the basis of the therapeutic strategy.4, 7, 8, 9 However, FIGO staging fails to identify patients with high risk of death who could potentially be eligible for chemotherapy.10, 11, 12, 13 Even if this disease is clinically very aggressive,5 it is difficult to predict the outcome especially when the diagnosis is made at stage I (confined to the uterus). Recently, a specific uterine leiomyosarcomas nomogram for predicting post-resection 5-year overall survival using seven clinicopathological items (age, tumor size, tumor grade, cervical involvement, loco-regional metastases, distant metastases and mitotic index) was published14 and was subsequently validated on an independent series.15 In this nomogram, the mitotic index and the tumor grade were taken into account as biological parameters. Nevertheless, the tumor grading in uterine leiomyosarcomas is controversial because by definition leiomyosarcomas diagnosed on the basis of Stanford criteria6 are of high grade. Contrary to other soft tissue sarcomas, the tumor grade does not show a prognostic value in uterine leiomyosarcomas.16 Among the seven parameters, three (presence of regional metastasis, distant metastasis and size) are strictly linked to the FIGO stage.14
Hence, there is a need to clarify these prognostic strategies. A few years ago, we published a new classification method based on genomic profiling complementary to the morphology able to distinguish within the STUMP category those uterine smooth muscle lesions with a risk of recurrence and poor outcomes from benign lesions.17
In this study, we analyzed the genomic profile by array-comparative genomic hybridization in a series of 77 uterine smooth muscle lesions (44 leiomyosarcomas, 14 STUMP and 19 leiomyomas) to validate the power of genomic index as a recurrence predictor. Furthermore, we set out to improve our previous results by studying a larger series with a broader follow-up and to identify overall survival prognostic factors in uterine leiomyosarcomas.
Materials and methods
Tumor Samples
Seventy-seven formalin-fixed and paraffin-embedded uterine smooth muscle lesions from 76 patients (for one patient, both the primary tumor and the recurrence were examined) were collected in France through the French sarcoma network (RRePS and GYN RRePS) (Institut Bergonié of Bordeaux, Centre Oscar Lambret of Lille, Hôpitaux Universitaires Lyon Sud, Centre JF LeClerc, Dijon, Hôpital Universitaire of Poitiers, Centre Jean Perrin, Clermont-Ferrand, Centre Alexis Vautrin, Vandouevre-les Nancy, Institut Gustave Roussy, ICO, Paul Papin, Angers), from Belgium (Hopitaux Universitaires de Bruxelles, Catholic University of Leuven), Czech Republic (University Hospital from Prague) and Switzerland (Hôpitaux Universitaires and Argot-Lab of Lausanne). Fourteen uterine tumors diagnosed as STUMP along with 19 uterine leiomyomas previously published17 were included in the series. The tumors were diagnosed between 1977 and 2013. For each patient, 1–8 slides were available (mean: 2 slides). All cases were centrally reviewed by one of the authors (SC) and classified according to Stanford criteria6 and 2014 WHO guidelines for female reproductive organs.2 Cytologic atypia was evaluated at medium power magnification (objective × 10) according to the presence of high nuclear size or nuclear pleomorphism and hyperchromatism,18 the mitotic count was evaluated on 10 high-power fields (objective × 40, field diameter 0.53 mm) and the tumor cell necrosis defined by an abrupt transition from necrotic to non-necrotic areas without interposed granulation tissue.18 The mitotic cut-off was 10 mitoses/10 power fields for spindle cell smooth muscle tumors, ≥4 mitoses for epithelioid and ≥2 mitoses for myxoid tumors.2, 18
Frozen material was available for five samples. The samples from the tumor archives of each participating department were centralized in the Biological Resources Center of Institut Bergonié, which the French authorities authorized for scientific research (AC-2008-812).
DNA and RNA Extraction
Genomic DNA was extracted from formalin-fixed and paraffin-embedded tissues according to the protocol for DNA isolation from formalin-fixed and paraffin-embedded tissues (http://www.chem-agilent.com/pdf/G441090020v3_1_CGH_ULS_Protocol.pdf) (Agilent Technologies, Santa Clara, CA, USA). A cut-off of 50% of cellularity in tumor samples was set for the analysis.
Array-Comparative Genomic Hybridization Analysis
DNA was hybridized onto 8 × 60 K whole-genome arrays (G4450A; Agilent Technologies) according to the manufacturer’s protocol. Microarray slides were scanned using a DNA Microarray Scanner, images were analyzed by Feature Extraction V10.1.1.1 followed by Agilent Cytogenomic software 4.0. The ADM-2 algorithm of the Comparative Genomic Hybridization Analytics v4.0.76 software (Agilent Technologies) was used to identify the DNA copy number anomalies at the probe level. A low-level copy number gain was defined as a log 2 ratio >0.25 and a copy number loss was defined as a log 2 ratio <0.25. A high-level gain or amplification was defined as a log 2 ratio >1.5 and a homozygous deletion was suspected when the ratio was <-1. The range for derivative log ratio spread cut-off was fixed to 0.50. Genomic index was calculated for each profile as follows: genomic index=A2/C, where A is the total number of alterations (segmental gains and losses) and C is the number of involved chromosomes.19, 17
Statistical Analysis
Metastasis-free survival was calculated by the Kaplan–Meier method from the date of initial diagnosis to the date of first metastasis or last follow-up. Overall survival, using the Kaplan–Meier method, was calculated from the date of diagnosis to death or last follow-up. Survival curves were compared with the log-rank test. Univariate survival analyses were performed by using the R software version 3.4.0 (R Development Core Team, Vienna, Austria, 2009) and the ‘survival’ package (A Package for Survival Analysis in S; Terry Therneau, February 2002; R package, version 2.40-1). To test whether gene alterations (losses or gains) are enriched in groups of tumors classified by the genomic index, Fisher's exact tests were performed. Multivariate survival analyses were performed using Cox regression with Firth’s correction with the ‘coxphf’ package (Georg Heinze and Meinhard Ploner, 2016, version 1.12).
Results
Pathologic Features
After centralized pathological review, the series comprised 19 leiomyomas, 14 STUMP and 44 leiomyosarcomas. Morphologically, all leiomyomas but one case of bizarre nuclei leiomyoma, showed conventional spindle cell morphology. Spindle cell features were observed in the STUMP group (all 14 tumors).
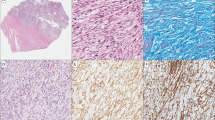
Among the 44 leiomyosarcomas (43 patients), 29/44 tumors showed a spindle cell morphology, 10/44 epithelioid, 4/44 pleomorphic with giant osteoclastic cells (with negative stains for melanocytic markers) and 1/44 myxoid (Figure 1). The clinicopathological data are summarized in Table 1.
A representation of the morphological features of uterine smooth muscle lesions. (a) Uterine leiomyosarcoma with spindle cells, pleomorphic and osteoclastic features and atypical mitoses, stage FIGO IB and genomic index=84. The patient developed lung metastases and died 32.4 months after the diagnosis (patient 68). (b) Myxoid leiomyosarcoma with infiltration of myometrial wall. Some areas are highly cellular with four mitoses, stage FIGO 2B and genomic index=13.5 (patient 47). (c) Epithelioid leiomyosarcoma with genomic index=27. The patient died of disease 26 months after the diagnosis with peritoneal metastases (patient 48). (d) Smooth muscle lesion with mild atypia, 20 mitoses (STUMP) with genomic index=14.3. The patient had multiple peritoneal and retroperitoneal recurrences and she is alive after 103.2 months (patient 33).
Genomic Data and Clinical Correlations
Genome complexity evaluation by genomic index assessment (quantitative approach) and prognostic value
Array-comparative genomic hybridization was analyzable in all 77 tumors. Follow-up data were available for all but two patients with leiomyomas (mean follow-up: 63.6 months, range: 9–232 months). The genomic profiling split the present series of uterine smooth muscle lesions in two groups according to the cut-off defined in our previous paper (genomic index=10):17 a group with genomic index <10 (19/74) and a second group with genomic index ≥10 (55/74; Figure 2a).
Genomic index and genomic alterations and clinical outcome. (a) Histogram. The number of patients with relapse (black bar) and without relapse (gray bar) shown in x axis; the genomic index level in y axis. The genomic index at the cut-off of 10 splits the population of 74 patients with uterine smooth muscle lesion in a group of 19 patients without metastases and a group of 55 patients with 44 metastatic events. (b) Kaplan-Meier relapse-free survival analysis of 74 uterine smooth muscle tumors according the genomic index at the cut-off of 10. (c) Kaplan-Meier for overall survival according to genomic index at the cut-off of 35 in the subgroup with genomic index ≥10 (genomic leiomyosarcomas). (d) Kaplan-Meier analysis of overall survival in the group with genomic index ≥10 (genomic leiomyosarcomas) with 5p gain. (e) Kaplan-Meier analysis of overall survival in the group with genomic index ≥10 (genomic leiomyosarcomas) with 5p gain with tumors limited to the uterus (stage I). (f) Kaplan-Meier analysis of overall survival in the group with genomic index ≥10 (genomic leiomyosarcomas) with tumors limited to the uterus (stage I) according to genomic index at the cut-off of 35.
The first group (genomic index <10) is characterized by a low level of chromosomal rearrangements (Figure 3a; mean genomic index: 2.3, range: 0–9.14) in contrast with the second group (genomic index ≥10) harboring complex genomic profiles (mean genomic index: 51.8, range: 11–180; Figure 3b).
Penetrance plots of the different subgroups classified according to genomic index. (a) Penetrance plot of the tumors with genomic index <10 (benign tumors: all leiomyomas and two STUMPs). (b) Penetrance plot of tumors with genomic index >10 (malignant tumors: all leiomyosarcomas and 12 STUMPs). (c) Penetrance plot of tumors with genomic index >10 (malignant tumors: all leiomyosarcomas and 12 STUMPs) of patients alive. (d) Penetrance plot of tumors with genomic index >10 (malignant tumors: all leiomyosarcomas and 12 STUMPs) of patients dead of disease.
The Kaplan–Meier analysis demonstrated a significant difference in clinical outcome with no recurrence in the group with genomic index <10 and recurrences and deaths in the second group (Figure 2b), confirming our previous results.17
Morphologically, the first group included all leiomyomas, two STUMPs and no leiomyosarcomas. The second group included all leiomyosarcomas and 12 STUMPs. Only four patients with leiomyosarcomas did not recur (of note, one patient died of pulmonary embolism 9 days after surgery) and among the 12 STUMPs with genomic index ≥10, seven recurred (Table 1).
All leiomyomas showed a flat or very simple profile (mean genomic index: 1.9, range: 0–9.1). The leiomyosarcomas group showed a complex, rearranged chromosome profile with numerous intra-chromosomal breaks (gains and losses). The mean genomic index in the leiomyosarcomas group was 55 (range: 13.5–180). The STUMP group was then split into two groups according to genomic index. Owing to the clinical aggressiveness and outcome of the tumors with genomic index ≥10, they were thereafter considered as leiomyosarcomas.
Therefore, the second step was to identify a death predictor in this newly defined group of leiomyosarcomas. Different genomic index cut-offs (five for each step) were tested and a cut-off of genomic index=35 was identified as an efficient predictor of death (P=0.035, HR=2.18 (1.04–4.58); Figure 2c).
In order to identify specific chromosomal alterations (qualitative approach) related to prognostic value, we analyzed differential penetrance plots among the patients alive, whose tumor had genomic index ≥10 (19/55) and patients who died of the disease (36/55). We identified five frequent alterations, in addition to genomic index ≥35 (Figure 2c), associated with overall survival: 5p gain (Figure 2d,Table 2a), 1p gain, 13q loss (including RB1) and 17p gain including MYOCD (Table 2a). Staging being the gold standard for clinical/pathological prognosis, all markers (including genomic index ≥35) were tested for independency against the stage in a multivariate analysis. Stage and 5p gain were shown to be statistically independent prognostic factors at multivariate analysis (stage: P<0.001, HR=4.36 (1.82–10.42), 5p gain: P=0.04, HR=3.24 (1.04–10.04); Table 2a). Given that stage I tumors can have a risk of metastasis, we then further refined staging by testing molecular prognostic factors within stage. Among the tumors with genomic index ≥10 and stage I (37/55 patients), four overall survival prognostic factors were still significant: 5p gain (P<0.001, HR=4.88 (1.74–13.7); Figure 2e), genomic index at the cut-off of 35 (P=0.0193, HR=3.2 (1.15–8.92); Figure 2f), 13 chromosome loss including RB1 (P=0.0096, HR=9.04 (1.2–67.81)) (Supplementary Figure 1A) and 17p gain including MYOCD (P=0.0425, HR=2.45 (1–5.97)) (Supplementary Figure 1B).
Among the other clinicomorphological parameters tested, the presence of moderate and marked atypia (Fisher’s P=0.043), the presence of tumor necrosis (Fisher’s P=0,001) and a mitotic index (cut-off ≥20; t-test P<0.001) were poor prognostic markers for overall survival (Table 2b,Supplementary Figure 1C). For these parameters, multivariate analysis showed that genomic index ≥35 remained significantly independent (P=0.0333; Table 2c).
Correlation Between Chromosomal Alterations and Morphology
No correlation was found between genomic index and morphology of the tumor (spindle, epithelioid or myxoid). No correlation was observed between any specific genomic alteration (5p gain, 17p loss, 13 loss, 17p gain) and tumor morphology (spindle vs epithelioid).
Discussion
In the last 5 years, many analyses based on whole-genome approaches have improved our knowledge of uterine smooth muscle lesion biology.20, 21, 22 Nevertheless, the routine diagnostic practice lacks complementary diagnostic tools. For uterine leiomyosarcomas, despite very aggressive clinical features,5 it is difficult to predict the outcome, especially when the diagnosis is made at stage I (tumor confined to the uterus).
According to the literature,8, 10, 23 adjuvant treatment of a stage I uterine leiomyosarcomas is an option without evidence of benefit. There is a need for clinical trials to highlight the benefit of chemotherapy in terms of overall survival and relapse-free survival. In absence of clinical prognostic markers, the identification of new genomic prognostic markers appears critical for setting up clinical trials aiming to evaluate new treatments in uterine leiomyosarcomas. Genomic prognostic markers could have an essential role in deciding on adjuvant chemotherapy for stage I uterine leiomyosarcomas. In our series of 37 molecular leiomyosarcomas (genomic index ≥10) at stage I, four genomic prognostic markers correctly separated the outcomes for patients alive or dead: 5p gain, chromosome 13 loss including RB1, proximal 17p gain with MYOCD and genomic index ≥35.
Further analyses are required to understand whether this is due to a chromosomal mechanism (specific or general) or due to genes located in these regions that are specifically overexpressed as a consequence of a chromosomal gain. The 5p gain was previously reported in extra-uterine24 and uterine leiomyosarcomas25 but no association with outcome was observed. The 17p proximal gain, including the MYOCD gene, was previously reported in literature in soft tissue leiomyosarcomas24, 26, 27, 28 and in uterine leiomyosarcomas.25 MYOCD gene induces smooth muscle differentiation and promotes cell migration.27 In a human uterine leiomyosarcomas cell line, MYOCD induced a phenotypic cell switch from a dedifferentiated to a differentiated smooth muscle phenotype.29 MYOCD expression level controls smooth muscle differentiation protein expression and has an impact on cell migration in soft tissue leiomyosarcomas.27 Furthermore, it confers aggressive outcome in soft tissue sarcomas.30
Hu et al25 found a gain of 17p in 38% of the uterine leiomyosarcomas and interestingly, no 17p gain was found in alive patients (4/19). Chromosome 13 was lost in 80% of the leiomyosarcomas in our series and is the most common genomic event in uterine (76%) 25 and extra-uterine leiomyosarcomas (ranging from 54% 28, 24 to 71% 26) and in the majority of the cases, correlation between this event and follow-up was not established.
The morphological analysis based on the presence of atypia, mitotic count and tumor cell necrosis correlated to a poor outcome in our series. The prognostic value of cytological atypia,31, 32 as well the mitotic count5, 33, 34 was reported in previous publications. However, atypia and mitoses could be difficult to assess and there is only a moderate inter-observer agreement on tumor cell necrosis in uterine smooth muscle lesions among gynecological pathologists.35 Genomic index assessment could be a useful tool, as highlighted in our multivariate analysis (Table 2c), to avoid such diagnostic discrepancies.
In our series, the genomic index cut-off of 10 splits the STUMP group into two: a flat or very simple genomic profile group akin to leiomyomas and a group of tumors with complex genomic profile similar to leiomyosarcomas (with recurrences and deaths) thereby erasing the STUMP category. In the benign lesions group with genomic index <10 (all leiomyomas and two STUMP), there were no chromosomal alterations such as RB1 and TP53 loss (Table 1). One exception is the bizarre nuclei leiomyoma case, with a borderline genomic index=9, which showed chromosome 13 loss including the RB1 gene and chromosome 17p loss including theTP53 gene. These alterations have already been reported in these benign lesions.36, 37, 38, 39 In fact, some bizarre nuclei leiomyomas inexplicably show rearranged profiles (in our experience lower than leiomyosarcomas) and a good outcome. Furthermore, the origin of this subtype of leiomyoma is not clear either.
Genomic profiling by array-comparative genomic hybridization on formalin-fixed and paraffin-embedded samples is a useful, easy and accessible tool complementary to the morphological approach. It is a diagnostic tool that splits the STUMP category into benign (leiomyoma) and malignant (leiomyosarcomas) tumors. It is a prognostic marker and a predictor of overall survival in stage I uterine leiomyosarcomas. Indeed, all the comparative genomic hybridization analyses on our series were performed on formalin-fixed and paraffin-embedded and -extracted DNA, with 100% feasibility. Genomic profiling could be used even on a limited amount of material such as pre-operative biopsies in order to guide surgical intervention (hysterectomy vs myomectomy or a minimally invasive surgery).
In conclusion, we have demonstrated that STUMP classification could be overcome by utilizing genomic index at the cut-off of 10. The 5p gain as genomic event and the stage as clinical parameter are poor overall survival prognostic factors in uterine leiomyosarcomas. In stage I tumors, the 5p gain, 17p gain, chromosome 13 loss and genomic index at the cut-off of 35 are poor prognostic factors of overall survival and therefore they could be potential parameters for randomization in prospective clinical trials. This approach opens the way to new insights into uterine and other gynecological smooth muscle lesions and would allow reclassification of lesions according to genomic complexity. In fact, there is a continuum gradient of genomic complexity and instability correlating with tumor aggressiveness. As genomic profiling by array-comparative genomic hybridization on formalin fixed and paraffin embedded is feasible and accessible in hospital laboratories, this approach could be used in routine practice as a complement to histology.
References
Skorstad M, Kent A, Lieng M . Uterine leiomyosarcoma - incidence, treatment, and the impact of morcellation. A nationwide cohort study. Acta Obstet Gynecol Scand 2016;95:984–990.
Oliva E, Carcangiu ML, Carinelli SG et al. Mesenchymal tumours. Smooth muscle tumour of uncertain malignant potential. In: Kurman RJ CM, Herrington CS, Young RH (eds). WHO Classification of Tumours of Female Reproductive Organs, 2014, pp 135–147.
Baird DD, Dunson DB, Hill MC et al. High cumulative incidence of uterine leiomyoma in black and white women: ultrasound evidence. Am J Obstet Gynecol 2003;188:100–107.
Ricci S, Stone RL, Fader AN . Uterine leiomyosarcoma: epidemiology, contemporary treatment strategies and the impact of uterine morcellation. Gynecol Oncol 2017;145:208–216.
Garcia C, Kubat JS, Fulton RS et al. Clinical outcomes and prognostic markers in uterine leiomyosarcoma: a population-based cohort. Int J Gynecol Cancer 2015;25:622–628.
Bell SW, Kempson RL, Hendrickson MR . Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol 1994;18:535–558.
Amant F, Coosemans A, Debiec-Rychter M et al. Clinical management of uterine sarcomas. Lancet Oncol 2009;10:1188–1198.
Hensley ML, Ishill N, Soslow R et al. Adjuvant gemcitabine plus docetaxel for completely resected stages I-IV high grade uterine leiomyosarcoma: results of a prospective study. Gynecol Oncol 2009;112:563–567.
Amant F, Lorusso D, Mustea A et al. Management strategies in advanced uterine leiomyosarcoma: focus on trabectedin. Sarcoma 2015;2015:704124.
Omura GA, Blessing JA, Major F et al. A randomized clinical trial of adjuvant adriamycin in uterine sarcomas: a Gynecologic Oncology Group Study. J Clin Oncol 1985;3:1240–1245.
Reed NS, Mangioni C, Malmstrom H et al. Phase III randomised study to evaluate the role of adjuvant pelvic radiotherapy in the treatment of uterine sarcomas stages I and II: an European Organisation for Research and Treatment of Cancer Gynaecological Cancer Group Study (protocol 55874). Eur J Cancer 2008;44:808–818.
Rose PG, Boutselis JG, Sachs L . Adjuvant therapy for stage I uterine sarcoma. Am J Obstet Gynecol 1987;156:660–662.
Sorbe B, Johansson B . Prophylactic pelvic irradiation as part of primary therapy in uterine sarcomas. Int J Oncol 2008;32:1111–1117.
Zivanovic O, Jacks LM, Iasonos A et al. A nomogram to predict postresection 5-year overall survival for patients with uterine leiomyosarcoma. Cancer 2012;118:660–669.
Iasonos A, Keung EZ, Zivanovic O et al. External validation of a prognostic nomogram for overall survival in women with uterine leiomyosarcoma. Cancer 2013;119:1816–1822.
Pautier P, Genestie C, Rey A et al. Analysis of clinicopathologic prognostic factors for 157 uterine sarcomas and evaluation of a grading score validated for soft tissue sarcoma. Cancer 2000;88:1425–1431.
Croce S, Ribeiro A, Brulard C et al. Uterine smooth muscle tumor analysis by comparative genomic hybridization: a useful diagnostic tool in challenging lesions. Mod Pathol 2015;28:1001–1010.
Oliva E . Practical issues in uterine pathology from banal to bewildering: the remarkable spectrum of smooth muscle neoplasia. Mod Pathol 2016;29 (Suppl 1):S104–S120.
Lagarde P, Perot G, Kauffmann A et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res 2012;18:826–838.
Mehine M, Kaasinen E, Makinen N et al. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med 2013;369:43–53.
Mehine M, Kaasinen E, Aaltonen LA . Chromothripsis in uterine leiomyomas. N Engl J Med 2013;369:2160–2161.
Makinen N, Aavikko M, Heikkinen T et al. Exome sequencing of uterine leiomyosarcomas identifies frequent mutations in TP53, ATRX, and MED12. PLoS Genet 2016;12:e1005850.
Hempling RE, Piver MS, Baker TR . Impact on progression-free survival of adjuvant cyclophosphamide, vincristine, doxorubicin (adriamycin), and dacarbazine (CYVADIC) chemotherapy for stage I uterine sarcoma. A prospective trial. Am J Clin Oncol 1995;18:282–286.
El-Rifai W, Sarlomo-Rikala M, Knuutila S et al. DNA copy number changes in development and progression in leiomyosarcomas of soft tissues. Am J Pathol 1998;153:985–990.
Hu J, Khanna V, Jones M et al. Genomic alterations in uterine leiomyosarcomas: potential markers for clinical diagnosis and prognosis. Genes Chromosomes Cancer 2001;31:117–124.
Larramendy ML, Kaur S, Svarvar C et al. Gene copy number profiling of soft-tissue leiomyosarcomas by array-comparative genomic hybridization. Cancer Genet Cytogenet 2006;169:94–101.
Perot G, Derre J, Coindre JM et al. Strong smooth muscle differentiation is dependent on myocardin gene amplification in most human retroperitoneal leiomyosarcomas. Cancer Res 2009;69:2269–2278.
Agaram NP, Zhang L, LeLoarer F et al. Targeted exome sequencing profiles genetic alterations in leiomyosarcoma. Genes Chromosomes Cancer 2016;55:124–130.
Kimura Y, Morita T, Hayashi K et al. Myocardin functions as an effective inducer of growth arrest and differentiation in human uterine leiomyosarcoma cells. Cancer Res 2010;70:501–511.
Perot G, Mendiboure J, Brouste V et al. Smooth muscle differentiation identifies two classes of poorly differentiated pleomorphic sarcomas with distinct outcome. Mod Pathol 2014;27:840–850.
Nordal RR, Kristensen GB, Kaern J et al. The prognostic significance of stage, tumor size, cellular atypia and DNA ploidy in uterine leiomyosarcoma. Acta Oncol 1995;34:797–802.
Wang WL, Soslow R, Hensley M et al. Histopathologic prognostic factors in stage I leiomyosarcoma of the uterus: a detailed analysis of 27 cases. Am J Surg Pathol 2011;35:522–529.
Pelmus M, Penault-Llorca F, Guillou L et al. Prognostic factors in early-stage leiomyosarcoma of the uterus. Int J Gynecol Cancer 2009;19:385–390.
Davidson B, Kjaereng ML, Forsund M et al. Progesterone receptor expression is an independent prognosticator in FIGO stage I uterine leiomyosarcoma. Am J Clin Pathol 2016;145:449–458.
Lim D, Alvarez T, Nucci MR et al. Interobserver variability in the interpretation of tumor cell necrosis in uterine leiomyosarcoma. Am J Surg Pathol 2013;37:650–658.
Downes KA, Hart WR . Bizarre leiomyomas of the uterus: a comprehensive pathologic study of 24 cases with long-term follow-up. Am J Surg Pathol 1997;21:1261–1270.
Croce S, Young RH, Oliva E . Uterine leiomyomas with bizarre nuclei: a clinicopathologic study of 59 cases. Am J Surg Pathol 2014;38:1330–1339.
Liegl-Atzwanger B, Heitzer E, Flicker K et al. Exploring chromosomal abnormalities and genetic changes in uterine smooth muscle tumors. Mod Pathol 2016;29:1262–1277.
Zhang Q, Ubago J, Li L et al. Molecular analyses of 6 different types of uterine smooth muscle tumors: emphasis in atypical leiomyoma. Cancer 2014;120:3165–3177.
Acknowledgements
This study was supported by the grants from ARC and la Ligue Régionale contre le cancer. We thank Dr Ravi Nookala of Institut Bergonié for the medical writing service and Magali Philip, Quitterie Fontanges, Patrick Murat and Virginie Duvignau for photographs. FA is senior researcher for the Research Fund Flanders (F.W.O.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Croce, S., Ducoulombier, A., Ribeiro, A. et al. Genome profiling is an efficient tool to avoid the STUMP classification of uterine smooth muscle lesions: a comprehensive array-genomic hybridization analysis of 77 tumors. Mod Pathol 31, 816–828 (2018). https://doi.org/10.1038/modpathol.2017.185
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2017.185
This article is cited by
-
Uterine leiomyosarcomas with osteoclast-like giant cells associated with high expression of RUNX2 and RANKL
Virchows Archiv (2021)
-
NFATc2-rearranged sarcomas: clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker
Modern Pathology (2020)
-
Molecular and clinicopathologic characterization of intravenous leiomyomatosis
Modern Pathology (2020)
-
Smooth muscle tumors of the gastrointestinal tract: an analysis of prognostic features in 407 cases
Modern Pathology (2020)
-
Myxoid smooth muscle neoplasia of the uterus: comprehensive analysis by next-generation sequencing and nucleic acid hybridization
Modern Pathology (2019)