Abstract
Translocation renal cell carcinoma and succinate dehydrogenase (SDH)-deficient renal cell carcinoma are now recognized as specific renal tumor types in the World Health Organization (WHO) classification. Both have limited immunohistochemical positivity for epithelial markers, and the spectrum of morphology continues to widen for both of these entities. We identified four renal cell carcinomas with positive TFE3 immunohistochemical staining and negative SDHB staining. The patients (2F, 2M) ranged in age from 19 to 65 years. All tumors were composed, at least in part, of eosinophilic cells. Cytoplasmic inclusions, prominent nucleoli, and mitotic figures were seen in three tumors. Psammoma bodies were also present in two tumors. Using immunohistochemistry, a broad spectrum of commonly used renal tumor markers yielded nonspecific, limited positivity, including uniformly positive reactions for PAX8 but negative results for cathepsin K and HMB45. Fluorescence in situ hybridization results showed the presence of TFE3 gene rearrangement in all four tumors, and molecular analysis revealed SDHB mutations in neoplastic cells of three tumors. In one case, the same SDHB mutation was confirmed in the adjacent non-neoplastic tissue. We report for the first time the presence of both TFE3 translocation and SDHB mutation in the same tumor.
Similar content being viewed by others
Main
Our understanding of the genetics of renal cell carcinoma has been evolving in the last three decades, with recognition of a number of new entities. In hereditary renal tumor syndromes and with translocation renal cell carcinomas, the tumors may present in unusually young patients, although they both also affect adults in the typical age range for renal cell carcinoma in general. Among these, the microphthalmia transcription factor (MITF) family of translocation renal cell carcinomas is well recognized,1 and more recently succinate dehydrogenase (SDH)-deficient renal cell carcinoma has been recognized.2, 3 Xp11 translocation renal cell carcinoma is cytogenetically characterized by chromosomal translocations involving breakpoints in the TFE3 gene, which maps to the Xp11.2 locus. Histologically, a wide spectrum of morphology has been described in these tumors, emphasizing the need to consider these carcinomas in the differential diagnosis of unusual renal cell carcinomas occurring in either children or adults.1 Strong nuclear TFE3 immunohistochemical expression is a reasonably sensitive and specific marker for Xp11 translocation renal cell carcinoma.4 However, fluorescence in situ hybridization (FISH) assays have been demonstrated to be more reliable.5
SDH-deficient renal cell carcinoma is now accepted as a specific tumor type in the World Health Organization classification,6 and is primarily thought to affect patients with germline mutations of SDH gene subunits. Germline mutations in one of the SDH subunit genes (A, B, C, or D) are associated with the hereditary paraganglioma/pheochromocytoma syndromes, as well as development of gastrointestinal stromal tumors and pituitary adenomas. Many of the reported SDH-deficient renal cell carcinomas to date have been microscopically characterized by sheets of uniform eosinophilic cells with flocculent cytoplasmic inclusions.2, 3 However, other morphologies have been reported.7 To identify these tumors, the absence of immunohistochemical staining for SDH subunit B (SDHB) is considered a reliable tool.2, 3, 8, 9, 10
Both translocation renal cell carcinoma and SDH-deficient renal cell carcinoma may occur in young patients, and their morphology may overlap with those of other types of renal cell carcinoma. When this occurs, immunostaining for TFE3 and SDHB can be helpful. In this study, we identified four renal cell carcinomas with strong nuclear TFE3 staining and absence of immunohistochemical staining for SDHB. We also demonstrated the presence of both TFE3 gene translocation and SDHB gene mutations in three of the tumors, and SDHB germline mutation in one of them.
Materials and methods
Patients and Samples
Four consultation cases of renal cell carcinoma with TFE3 overexpression and immunohistochemical loss of SDHB were retrieved from the Department of Pathology of Indiana University. None of these cases was previously reported. Hematoxylin and eosin stained-slides and paraffin blocks were available for each case. This research was approved by the Institutional Review Board.
Immunohistochemistry
Immunohistochemical analysis was performed on all cases utilizing the SDHB monoclonal antibody (clone 21A11, Abcam; dilution 1:200), on whole tissue sections. Granular cytoplasmic staining was considered as positive. Complete absence of staining in the neoplastic cells in the presence of positive internal control reactions in non-neoplastic tissue was considered to be a negative reaction.
An immunohistochemical panel to assess the likelihood of translocation-associated renal cell carcinoma was used: antibodies to cathepsin K (monoclonal mouse antihuman antibody, 3F9; Abcam; dilution 1:800), HMB45 (monoclonal mouse antihuman antibody, HMB45; DAKO; prediluted), melan-A (monoclonal mouse antihuman antibody, A103; DAKO; prediluted), and TFE3 (monoclonal mouse antihuman antibody, MRQ37, Cell Marque; dilution 1:500). TFE3 immunohistochemistry was performed using the DAKO’s Autostainer Plus machine.
In addition, the following immunohistochemical markers were used: antibodies to PAX8 (rabbit polyclonal antibody, Cell Marque; prediluted), vimentin (monoclonal mouse antihuman antibody, V9; DAKO; prediluted) CD10 (monoclonal mouse antihuman antibody, 56C6; DAKO; prediluted), AMACR (monoclonal rabbit antihuman P504S, 13H4 clone, Dako; prediluted), cytokeratin AE1/AE3 (monoclonal mouse antihuman antibody, AE1/AE3, Dako; prediluted), EMA (monoclonal mouse antihuman antibody, E29, Dako; prediluted), CD117 (rabbit monoclonal antibody, EP10, Cell Marque; prediluted), cytokeratin 7 (monoclonal mouse antihuman antibody, OV-TL 12/30, Dako; prediluted), and carbonic anhydrase IX (rabbit monoclonal antibody, EP161, Cell Marque; prediluted). Positive and negative controls yielded appropriate results for each antibody.
Fluorescence In Situ Hybridization
Interphase FISH assay was performed on all tumors as described previously.11 Tissue sections 4 μm thick were prepared from buffered formalin-fixed, paraffin-embedded tissue blocks containing tumor. The slides were deparaffinized with two washes with xylene (15 min each), and subsequently washed twice with absolute ethanol (10 min each), and then air dried in the hood. Then, the slides were treated with 10 mm citric acid (pH 6) (Zymed, San Francisco, CA, USA) at 95 °C for 10 min, rinsed in distilled water for 3 min, and then washed with 2 × SSC for 5 min. Digestion of the tissue was performed by applying 0.4 ml of pepsin (5 mg/ml in 0.1 m HCl/0.9 m NaCl) (Sigma, St Louis, MO, USA) at 37 °C for 40 min. The slides were rinsed with distilled water for 3 min, washed with 2 × SSC for 5 min, and air dried. The split-apart probe set for TFE3 uses BAC clones RP11-528A24 (116 kbp, located centromeric to TFE3, labeled with 5-fluorescein dUTP) and RP11-416B14 (182 kbp, located telomeric to TFE3, labeled with 5-ROX dUTP) (Empire Genomics, Buffalo, NY, USA). BAC clones for TFE3 were diluted with DenHyb2 at a ratio of 1:25. Five microlitres of diluted probe was applied to each slide in reduced light conditions. The slides were then covered with a 22 × 22-mm coverslip and sealed with rubber cement. Denaturation was achieved by incubating the slides at 83 °C for 12 min in a humidified box and hybridization at 37 °C overnight. The coverslips were removed, and the slides were washed twice with 0.1 × SSC/1.5 m urea at 45 °C (20 min each), and then washed with 2 × SSC for 20 min and with 2 × SSC/0.1% NP-40 for 10 min at 45 °C. The slides were further washed with room temperature 2 × SSC for 5 min. The slides were air dried and counterstained with 10 μl of 4′,6-diamidino-2-phenylindole (Insitus), covered with coverslips, and sealed with nail polish.
The slides were examined with a Zeiss Axioplan 2 microscope (Zeiss, Göttingen, Germany). The images were acquired with a CMOS camera, and analyzed with metasystem software (MetaSystem, Belmont, MA, USA). Five sequential focus stacks with 0.4-mm intervals were acquired and then integrated into a single image to reduce thickness-related artifacts; the methodology and analysis for this have been previously described.11 For each case, a minimum of 100 tumor cell nuclei were examined with fluorescence microscopy at × 1000 magnification. Only non-overlapping tumor nuclei were evaluated. The TFE3 fusion resulted in a split-signal pattern. Signals were considered to be split when the green and red signals were separated by a distance of two or more signal diameters. On this basis and based on other commercially available break-apart FISH assays and TFE3 break-apart FISH assays, a positive result was reported when ≥10% of the tumor nuclei showed the split-signal pattern.11
In our previous test validation, 100 neoplastic nuclei in 18 renal cell carcinomas with Xp11.2 translocation and 18 clear cell renal cell carcinomas were evaluated. The percentage of split signals ranged from 17 to 78% (mean 33%) in tumors with Xp11.2 translocation. In clear cell renal cell carcinomas (control), the percentage of split signals ranged from 0 to 7% (mean 2%).
SDHB Mutation Detection
A SDHB sequencing was performed using a capture-based next-generation sequencing panel on an Illumina HiSeq 2500 with libraries prepared from genomic DNA extracted from formalin-fixed paraffin-embedded tumor or non-neoplastic adjacent tissue.12 The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using open source and licensed software packages including BWA: 0.7.10-r789, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: 2014.4-3.3.0-0-ga3711, CNVkit: 0.3.3, Pindel, SATK: 2013.1-10- gd6fa6c3, Annovar: v2015Mar22, Freebayes, and Delly.
Results
Clinical Characteristics
The four cases included two women and two men. The ages ranged from 19 to 65 years old (mean 40 years). Two patients underwent radical nephrectomy and two patients underwent partial nephrectomy (three in left, one in right kidney).
Pathological Findings
The gross and microscopic features were different in each case and they are presented separately for completeness. A summary of the main histological findings is tabulated in Table 1 (Figures 1 and 2).
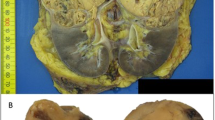
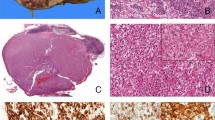
At low magnification, tumor 1 was partly surrounded by pseudocapsule and showed a solid architecture (a). At high magnification, the same tumor was composed of eosinophilic cells with central round nuclei and prominent nucleoli (c and d) with scattered foamy macrophages (c). Granular cytoplasmic SDHB staining was present in the non-neoplastic tubules (internal control) but all of the neoplastic cells were negative (b). Intense nuclear labeling for TFE3 was also observed (e). Fluorescence in situ hybridization of the same case showed the green and red signals split apart, demonstrating the TFE3 translocation (f).
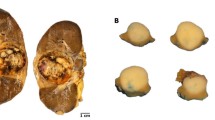
Morphologically, sheets of cells with eosinophilic cytoplasm (a) and cytoplasmic inclusion (b) were commonly seen. Some tumors displayed papillary architecture (c) and cystic changes (d). Aggregates of lymphocytes intermixed with tumor cells were observed in two tumors (e). In tumor 3, foci of metaplastic bone were found (f).
Case 1 (27-year-old woman)
A well-circumscribed mass was located in the superior-central left kidney. The lesion measured 93 × 77 × 68 mm and was described as tan-yellow with focal hemorrhage. Histologically, a pseudocapsule was present. The tumor was mainly composed of sheets or nests of polygonal cells with indistinct cell borders, abundant eosinophilic cytoplasm, central round nuclei, and prominent nucleoli. Prominent pale cytoplasmic inclusions were present. Mitotic figures were easily found (6 per 10 HPF). Entrapped non-neoplastic renal tubules and foamy macrophages were focally present. The pathological stage (2010) was pT2a pN0.
Case 2 (48-year-old man)
Macroscopically, a 37 × 34 × 30 mm tan-yellow mass was present in the left kidney. Histologically, the tumor was surrounded by a thick fibrous pseudocapsule. Entrapped renal tubules were occasionally seen at the periphery of the neoplasm. The tumor was mainly composed of sheets of cells with granular cytoplasm and round nuclei with pinpoint nucleoli and aggregates of intratumoral lymphocytes. Tubulopapillary architecture, formed by cells with eosinophilic cytoplasmic inclusions, round nuclei and prominent nucleoli, comprised the remainder of the tumor. A few psammoma bodies were seen. Sarcomatoid areas were also present. The tumor displayed extensive necrosis and hemorrhage. A hilar lymph node metastasis was present (1/1). The pathological stage (2010) was pT1a pN1.
Case 3 (65-year-old woman)
Macroscopically, the lesion measured 70 × 55 × 45 mm and was described as an ill-defined, pink-tan, and partially calcified mass with hemorrhagic areas. The tumor was partially delimited by a pseudocapsule. Architecturally, solid areas, tubular and papillary patterns, cystic changes, and metaplastic bone were observed. The carcinoma cells varied from uniform cells with pale eosinophilic cytoplasm, irregular nuclei and prominent nucleoli to polygonal cells with finely granular eosinophilic cytoplasm and enlarged hyperchromatic nuclei. Mitotic figures were occasionally encountered. Aggregates of intratumoral lymphocytes were found at the periphery of the tumor. Hemorrhage and necrosis were also present. The pathological stage (2010) was pT1b NX.
Case 4 (19-year-old man)
Macroscopically, an 18 × 17 mm lesion was observed in the right kidney and was described as a tan-orange nodule. Microscopically, the tumor was a well-circumscribed but unencapsulated nodule, made up of cells with abundant granular eosinophilic cytoplasm and distinct cell borders, arranged in small nests and tubules with focal cystic changes. The cells had uniform round nuclei with open chromatin and indistinct nucleoli. Mitotic figures were frequent (7 per 10 HPF). Cytoplasmic inclusions containing pale eosinophilic material and occasional small calcifications were present. The pathological stage (2010) was pT1a NX.
Immunohistochemical and Molecular Findings
Immunophenotypical and molecular features for each tumor are detailed in Table 2. Each of the four tumors showed strong and diffuse nuclear TFE3 immunolabeling and abnormal negative SDHB staining in the neoplastic cells. Strong and diffuse nuclear expression for PAX8 was consistently present. Staining for cathepsin K and HMB45 were not detected in any of the tumors. Staining for melan-A focally labeled neoplastic cells of tumor #2. Labeling for vimentin, epithelial membrane antigen and alpha-methylacyl-CoA racemase were present in all tumors, but varied from focal to diffuse positivity. Three of the four tumors labeled for cytokeratin AE1/AE3. Reactivity for CK7 was absent in all tumors. Carbonic anhydrase IX and CD10 varied from absent to patchy and diffuse positivity. All tumors but one showed negative staining for CD117.
FISH analysis demonstrated TFE3 gene translocation in the neoplastic cells of all four specimens. Split signals, indicating the TFE3 translocation, were found in 19% of tumor cells in tumors #1 and #2, in 12% of tumor cells in tumor #3 and in 14% of tumor cells in tumor #4.
Molecular analysis performed in the tumor tissue revealed splice site mutations in SDHB in tumors #1 and #4, and a missense mutation in tumor #3. No mutation in SDHB was identified in tumor #2. Two cases (cases #2 and #4) had sufficient normal tissue for SDHB mutation detection. The same SDHB mutation was found in normal adjacent tissue in case #4, suggesting germline SDHB mutation in this case. No SDHB mutation was found in tumor or adjacent normal tissue of case #2.
Discussion
SDH-deficient renal cell carcinoma is now recognized as a specific category of renal cell carcinoma in the 2016 WHO classification.6 The morphological features described to date as most helpful to identify this subtype of renal cell carcinoma include sheets of uniform cells with bubbly eosinophilic cytoplasm and intracytoplasmic vacuoles or inclusions (likely representing large abnormal mitochondria observed by electron microscopy).3, 9, 13 As experience with this tumor type is limited and its morphology may overlap with that of other types of renal cell carcinomas, it has recently been recommended that SDHB immunohistochemical staining be performed where there is clinical suspicion of familial renal cancer, regardless of the morphology.9 The range in age of patients with SDH-deficient renal cell carcinoma and germline SDH mutation is also broad (from 24 to 73 years).14 Therefore, the diagnosis, as well as that of translocation carcinoma, must be considered in not only young patients but also older adults. In addition, translocation renal cell carcinoma is increasingly recognized now as a consideration for the differential diagnosis of any unclassified or difficult to classify renal cell carcinoma, due to the broad spectrum of morphology that has been reported with these translocations. Papillary, solid, and nested architectures have been described. Prominent nucleoli, psammoma bodies, and necrosis are often present, whereas tumor lymphocytic aggregates and foamy macrophages are rarely observed.1 A variety of unusual patterns have been recently recognized in translocation carcinomas, including multilocular cystic tumors, oncocytic tumors, highly pleomorphic cells, and tumors mimicking urothelial carcinoma.5, 11
In the current study, we describe four renal cell carcinomas (2F, 2M; mean age=40 years) with positive immunohistochemical nuclear staining for TFE3 and abnormal immunohistochemical absence of SDHB. These were composed of sheets and nests of cells with tubules, papillae, and microcystic areas. As already mentioned, the variety of morphologic patterns and the presence of psammoma bodies are features frequently exhibited by translocation renal cell carcinomas.1 However, cystic changes2 as well as metaplastic bone formation,3 observed in case #3, have been reported in SDH-deficient renal cell carcinoma. Three tumors showed pale eosinophilic cytoplasmic inclusions, a distinct finding often seen in SDH-deficient renal cell carcinoma. Entrapped non-neoplastic renal tubules, similar to those reported in SDH-deficient renal cell carcinoma, were found in two of the tumors. Tumor lymphocytic aggregates throughout the tumor were observed in two other cases. Necrosis and hemorrhage were present in two tumors. In three tumors, mitotic figures were easily encountered, and one tumor displayed focally sarcomatoid features. These characteristics have been previously observed in translocation renal cell carcinomas and aggressive SDH-deficient renal cell carcinomas.15, 16
We demonstrated that all of the tumors with TFE3 nuclear staining showed TFE3 translocation using FISH. Although the percentage of cells with the TFE3 FISH split-signal pattern was low (12–19% of nuclei), it has been reported that a low percentage of split-signal nuclei may be observed in translocation-associated tumors when FISH is performed in paraffin-embedded tissue sections.5 Three of the four renal cell carcinomas with concomitant abnormal negative staining for SDHB had SDHB gene mutations in the carcinoma cells. The two splice site mutations (c.423+1G>A in case #1 and c.72+1G>T in case #4) have been already reported in two renal cell carcinomas with SDHB germline mutations.8, 13 We describe for the first time the occurrence of missense mutation SDHB p.V140F (case #3) in renal cell carcinoma. However, this mutation is not novel, as it has previously been found in paraganglioma/pheochromocytoma.17, 18, 19 Despite the absence of SDH immunohistochemical expression in tumor #2, molecular analysis failed to demonstrate a corresponding mutation in the SDHB gene. This finding could be explained by false negative staining, or as the result of mutation in another subunit of SDH. Loss of SDHB staining can result from disruption of the mitochondrial enzyme due to mutation in other SDH subunits, due to destabilization of the complex.20 Since alterations involving SDHA,15, 16 SDHC,7, 21, 22, 23 and SDHD7 genes are extremely rare, in this study we analyzed only SDHB, we cannot exclude the presence of another SDH mutation in case #2. On the other hand, abnormally absent or very weak SDHB immunolabeling in the absence of SDH mutation has been rarely reported in renal cell carcinoma, especially in cells with clear cytoplasm.9 In paraganglioma/pheochromocytoma, SDHB staining has been extensively studied and the false negative rate ranged from 9 to 16%.24, 25, 26 Of note, two of these studies used polyclonal rabbit antibody to SDHB,24, 26 and SDHA mutation was not investigated. Therefore, the absence of SDHB expression as the result of SDHA or other mutation remains a possibility.25, 26
So far, the majority of renal cell carcinomas with SDHB-negative staining appear to be associated with germline mutation of one of the SDH subunits.8, 9 However, somatic mutation in the SDHB gene has been observed in sporadic renal cell carcinoma. In 80 clear cell renal cell carcinomas studied by Papathomas et al, one tumor showed a heterogeneous pattern of SDHB staining, with abnormal negative staining in areas with sarcomatoid dedifferentiation and retained normal immunolabeling in low-grade tumor areas. Sequencing analysis demonstrated no germline mutation, but large intragenic SDHD and SDHAF2 deletions were found.27 In two cases (cases #2 and #4), SDHB mutational analysis was performed in the normal tissue adjacent to tumor. As expected, in case #2, no SDHB mutation was found. The same SDHB splice site mutation observed in tumor #4 was found in the adjacent normal tissue, and we considered it as an instance of SDH-deficient renal cell carcinoma occurring in association with germline mutation. In the two remaining cases, sufficient normal tissue was not available for evaluation.
Several speculations may be drawn about the coexistence of TFE3 translocation and SDHB mutation in renal cell carcinomas. One of the possibilities is that TFE3 translocation was the driver alteration in tumorigenesis and SDHB mutation occurred as a second mutation or vice versa. The second alteration might be the result of genomic instability or even have a role in tumor progression. Intriguingly, we observed aggressive features, such as sarcomatoid differentiation and mitotic figures in these tumors. Nevertheless, we acknowledge that double-hit SDH inactivation in the sporadic setting appears to be extremely rare and seldom reported in paraganglioma and pituitary adenoma.28, 29, 30 Conversely, whether TFE3 translocation may occur in patients with SDH germline mutation is unknown. By using immunohistochemistry, TFE3 expression has been investigated in a handful of SDH-deficient renal cell carcinomas and all of the seven cases studied were negative.3 Although the presence of germline mutation was found in only one case, the likelihood of SDH mutation as a driver in tumorigenesis in which TFE3 translocation subsequently occurs seems more likely. As this is the first report of this occurrence, concomitant investigations of these molecular abnormalities may be indicated in the work up of difficult or unusual renal cell carcinoma cases. A further comprehensive analysis of a large series is needed to address the biology and clinical behavior of these unusual tumors.
Of note, TFE3 translocation has recently been reported in renal cell carcinoma occurring in patients with prior neuroblastoma treated by chemotherapy.31 This association raises the question of whether TFE3 translocations may be found as a secondary event in other contexts and tumor types.
In conclusion, we describe the clinical and pathological characteristics of four renal cell carcinomas with TFE3 nuclear staining and negative SDHB immunolabeling. All tumors harbored TFE3 translocation and, in three of them, SDHB mutation was found.
References
Argani P . MiT family translocation renal cell carcinoma. Semin Diagn Pathol 2015; 32: 103–113.
Gill AJ, Hes O, Papathomas T et al, Succinate dehydrogenase (SDH)-deficient renal carcinoma: a morphologically distinct entity: a clinicopathologic series of 36 tumors from 27 patients. Am J Surg Pathol 2014; 38: 1588–1602.
Williamson SR, Eble JN, Amin MB et al, Succinate dehydrogenase-deficient renal cell carcinoma: detailed characterization of 11 tumors defining a unique subtype of renal cell carcinoma. Mod Pathol 2015; 28: 80–94.
Argani P, Lal P, Hutchinson B et al, Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol 2003; 27: 750–761.
Green WM, Yonescu R, Morsberger L et al, Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol 2013; 37: 1150–1163.
Moch H, Humphrey PA, Ulbright TM, Reuter VE . WHO classification of tumours of the urinary system and male genital organ. 4th edn, International Agency for Research on Cancer (IARC) Press: Lyon, France, 2016.
Ricketts CJ, Shuch B, Vocke CD et al, Succinate dehydrogenase kidney cancer: an aggressive example of the Warburg effect in cancer. J Urol 2012; 188: 2063–2071.
Gill AJ, Pachter NS, Clarkson A et al, Renal tumors and hereditary pheochromocytoma-paraganglioma syndrome type 4. N Engl J Med 2011; 364: 885–886.
Gill AJ, Pachter NS, Chou A et al, Renal tumors associated with germline SDHB mutation show distinctive morphology. Am J Surg Pathol 2011; 35: 1578–1585.
Miettinen M, Sarlomo-Rikala M, McCue P et al, Mapping of succinate dehydrogenase losses in 2258 epithelial neoplasms. Appl Immunohistochem Mol Morphol 2014; 22: 31–36.
Rao Q, Williamson SR, Zhang S et al, TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol 2013; 37: 804–815.
Joseph NM, Solomon DA, Frizzell N et al, Morphology and immunohistochemistry for 2SC and FH aid in detection of fumarate hydratase gene aberrations in uterine leiomyomas from young patients. Am J Surg Pathol 2015; 39: 1529–1539.
Housley SL, Lindsay RS, Young B et al, Renal carcinoma with giant mitochondria associated with germ-line mutation and somatic loss of the succinate dehydrogenase B gene. Histopathology 2010; 56: 405–408.
Ricketts C, Woodward ER, Killick P et al, Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst 2008; 100: 1260–1262.
Ozluk Y, Taheri D, Matoso A et al, Renal carcinoma associated with a novel succinate dehydrogenase A mutation: a case report and review of literature of a rare subtype of renal carcinoma. Hum Pathol 2015; 46: 1951–1955.
Yakirevich E, Ali SM, Mega A et al, A Novel SDHA-deficient renal cell carcinoma revealed by comprehensive genomic profiling. Am J Surg Pathol 2015; 39: 858–863.
Schimke RN, Collins DL, Stolle CA . Paraganglioma, neuroblastoma, and a SDHB mutation: resolution of a 30-year-old mystery. Am J Med Genet A 2010; 152A: 1531–1535.
Timmers HJ, Pacak K, Huynh TT et al, Biochemically silent abdominal paragangliomas in patients with mutations in the succinate dehydrogenase subunit B gene. J Clin Endocrinol Metab 2008; 93: 4826–4832.
Brouwers FM, Eisenhofer G, Tao JJ et al, High frequency of SDHB germline mutations in patients with malignant catecholamine-producing paragangliomas: implications for genetic testing. J Clin Endocrinol Metab 2006; 91: 4505–4509.
Barletta JA, Hornick JL . Succinate dehydrogenase-deficient tumors: diagnostic advances and clinical implications. Adv Anat Pathol 2012; 19: 193–203.
Malinoc A, Sullivan M, Wiech T et al, Biallelic inactivation of the SDHC gene in renal carcinoma associated with paraganglioma syndrome type 3. Endocr Relat Cancer 2012; 19: 283–290.
Gill AJ, Lipton L, Taylor J et al, Germline SDHC mutation presenting as recurrent SDH deficient GIST and renal carcinoma. Pathology 2013; 45: 689–691.
Shuch B, Agochukwu N, Ricketts CJ et al, Vascular endothelial growth factor receptor-targeted therapy in succinate dehydrogenase C kidney cancer. J Clin Oncol 2016; 34: e76–e79.
Papathomas TG, Oudijk L, Persu A et al, SDHB/SDHA immunohistochemistry in pheochromocytomas and paragangliomas: a multicenter interobserver variation analysis using virtual microscopy: a multinational study of the European network for the study of adrenal tumors (ENS@T). Mod Pathol 2015; 28: 807–821.
van Nederveen FH, Gaal J, Favier J et al, An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncol 2009; 10: 764–771.
Castelblanco E, Santacana M, Valls J et al, Usefulness of negative and weak-diffuse pattern of SDHB immunostaining in assessment of SDH mutations in paragangliomas and pheochromocytomas. Endocr Pathol 2013; 24: 199–205.
Papathomas TG, Gaal J, Corssmit EP et al, Non-pheochromocytoma (PCC)/paraganglioma (PGL) tumors in patients with succinate dehydrogenase-related PCC-PGL syndromes: a clinicopathological and molecular analysis. Eur J Endocrinol 2014; 170: 1–12.
Gill AJ, Toon CW, Clarkson A et al, Succinate dehydrogenase deficiency is rare in pituitary adenomas. Am J Surg Pathol 2014; 38: 560–566.
Gimm O, Armanios M, Dziema H et al, Somatic and occult germ-line mutations in SDHD, a mitochondrial complex II gene, in nonfamilial pheochromocytoma. Cancer Res 2000; 60: 6822–6825.
van Nederveen FH, Korpershoek E, Lenders JW et al, Somatic SDHB mutation in an extraadrenal pheochromocytoma. N Engl J Med 2007; 357: 306–308.
Falzarano SM, McKenney JK, Montironi R et al, Renal cell carcinoma occurring in patients with prior neuroblastoma: a heterogenous group of neoplasms. Am J Surg Pathol 2016; 40: 989–997.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Calió, A., Grignon, D., Stohr, B. et al. Renal cell carcinoma with TFE3 translocation and succinate dehydrogenase B mutation. Mod Pathol 30, 407–415 (2017). https://doi.org/10.1038/modpathol.2016.200
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.200
This article is cited by
-
Nuclear translocation of ASPL-TFE3 fusion protein creates favorable metabolism by mediating autophagy in translocation renal cell carcinoma
Oncogene (2021)
-
Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma
Nature Communications (2020)
-
NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma
Modern Pathology (2019)
-
RNA sequencing of Xp11 translocation-associated cancers reveals novel gene fusions and distinctive clinicopathologic correlations
Modern Pathology (2018)
-
Renal cell tumors with clear cell histology and intact VHL and chromosome 3p: a histological review of tumors from the Cancer Genome Atlas database
Modern Pathology (2017)