Abstract
Epigenetic alterations are hallmarks of cancer and powerful biomarkers, whose clinical utilization is made difficult by the absence of standardization and of common methods of data interpretation. The coordinate methylation of many loci in cancer is defined as ‘CpG island methylator phenotype’ (CIMP) and identifies clinically distinct groups of patients. In neuroblastoma (NB), CIMP is defined by a methylation signature, which includes different loci, but its predictive power on outcome is entirely recapitulated by the PCDHB cluster only. We have developed a robust and cost-effective pyrosequencing-based assay that could facilitate the clinical application of CIMP in NB. This assay permits the unbiased simultaneous amplification and sequencing of 17 out of 19 genes of the PCDHB cluster for quantitative methylation analysis, taking into account all the sequence variations. As some of these variations were at CpG doublets, we bypassed the data interpretation conducted by the methylation analysis software to assign the corrected methylation value at these sites. The final result of the assay is the mean methylation level of 17 gene fragments in the protocadherin B cluster (PCDHB) cluster. We have utilized this assay to compare the methylation levels of the PCDHB cluster between high-risk and very low-risk NB patients, confirming the predictive value of CIMP. Our results demonstrate that the pyrosequencing-based assay herein described is a powerful instrument for the analysis of this gene cluster that may simplify the data comparison between different laboratories and, in perspective, could facilitate its clinical application. Furthermore, our results demonstrate that, in principle, pyrosequencing can be efficiently utilized for the methylation analysis of gene clusters with high internal homologies.
Similar content being viewed by others
Main
Aberrant DNA methylation is considered an early event of cancer development and tumor progression.1, 2, 3 This epigenetic alteration is present, with different patterns of involved genes, in all type of cancer and is often, but not always, implicated in the functional inactivation of aberrantly methylated genes.
In 1999 Toyota et al.4 introduced the concept of CpG island methylator phenotype (CIMP), defined as the concordant methylation of multiple loci in cancer. CIMP, originally described in colorectal cancer, was later observed in other tumors and found to identify clinically distinct subgroups of patients.5, 6
In neuroblastoma (NB), a solid tumor of infancy, specific methylation biomarkers predictive of prognosis, or potentially useful to better classify the patients into risk groups, were identified and contributed to the definition of the CIMP in this tumor.7, 8, 9, 10
The protocadherin B cluster (PCDHB) includes genes that concur to define the methylator phenotype in NB. Because of the strong correlation between PCDHB methylation and patients’ survival, this cluster is considered the most informative member of CIMP in NB, and its methylation essentially recapitulates the entire predictive power of the multigenic CIMP.7 The PCDHB cluster seems to be a promising target of aberrant methylation also in breast cancer and Wilms tumor, but its analysis needs further investigations.11, 12
DNA methylation holds a highly promising role as a biomarker in the NB.10, 13, 14, 15, 16, 17 However, a comprehensive revision of the literature on the utilization of DNA methylation markers in the clinical practice shows that the absence of standardized technologies and of common methods of data interpretation makes very difficult the comparison of the results coming from different studies.18 As a consequence, until now, no methylation-based assay has entered in the routine clinical practice.
Recently, some tentative recommendations focused on the standardization of methods, type of biological samples and on the creation of databases to share data, and results were drawn as a prerequisite for the utilization of DNA methylation as a cancer biomarker. In particular, the quantitative detection of DNA methylation was considered essential to define CIMP in cancer.5, 18, 19
Along this line, we have developed a test on the basis of pyrosequencing, the gold standard technology for the quantitative detection of DNA methylation, to determine simultaneously the absolute methylation level of 17 members of the PCDHB cluster.
Our assay confirmed the prognostic value of the PCDHB cluster methylation, indicating the potential clinical usefulness of this biomarker in the NB.
Furthermore, the robustness and reproducibility of the pyrosequencing technique makes this approach ideal for the comparison of the results generated in different laboratories, and in perspective, this would facilitate the transfer of PCDHB methylation analysis into the clinical setting.
MATERIALS AND METHODS
Ethics Statement
The Ethics Committee of the Giannina Gaslini Children Hospital of Genova approved the collection, the storage in the Neuroblastoma Tissue Bank and the utilization of tumor samples. Informed consent was obtained from all patients or their relatives.
Patients
The analysis was conducted in two clinically distinct groups of NB patients classified at very low risk and high risk according to the criteria defined by the International Neuroblastoma Risk Group (INRG) Classification System.20 The very low-risk group included 19 patients at INSS21 stage 1, all alive 5 years after diagnosis. The high-risk group was composed of 18 patients at INSS stage 4, who died within 2 years from diagnosis. The clinical features of the patients are reported in Supplementary Table 1.
Extraction and Bisulfite Modification of Tumor DNA
The tumor DNA was provided by the Italian Neuroblastoma Tissue Bank.22 The specimens were collected at the onset of the disease and before therapy. A pathologist examined the tumor tissue utilized for nucleic acid extraction to verify the identity and homogeneity of the samples. The tumor cell content was at least 80%. DNA was isolated by proteinase K digestion and phenol/phenol–chloroform extraction.
DNA was modified with sodium bisulfite to convert the unmethylated Cs (but not the methylated Cs) into Ts with the Epitect Bisulfite kit of Qiagen (Milano, Italy) following the manufacturer instructions. Bisulfite-modified DNA of 60 ng was used to amplify the target sequences.
Pyrosequencing Assay of the PCDHB Cluster
The pyrosequencing assay23 was performed with a SPQ 96MA instrument (Qiagen). The primers were designed with the Pyrosequencing Assay Design Software (Qiagen) to recognize part of the CpG islands of 17 out of the 19 members of the PCDHB cluster. The three sequencing primers interrogated an average of 20 CpG sites (Table 1).
Sequencing reactions were performed with the Pyro Gold reagent kit SPQ 96MA, according to the manufacturer instructions. The sequencing analysis was conducted with the Pyro Q-CpG software (version 1.0.9) that also provides an internal control for the completeness of the bisulfite modification. The samples utilized in the present study passed this quality control.
MSqPCR Analysis
The two calibration curves for the methylated and unmethylated target were constructed with completely methylated and unmethylated DNA purchased from Qiagen. Methylation index was calculated as the fraction of methylated molecules in the total methylated and unmethylated DNA, utilizing the experimental conditions described by Abe et al.7
Statistical Analysis
The mean methylation value of the CpG doublets of the target sequences was considered for statistical analysis.
The correlation of the percentage of methylation between different parts of the highly homologous target fragment present in the 17 genes of the PCDHB cluster was assessed by computing the Pearson's linear correlation coefficient. The statistical differences in the methylation levels between very low- and high-risk NB patients were determined by the Student's t-test.
RESULTS
Pyrosequencing Assay Design of the PCDHB Cluster
The PCDHB cluster, mapping on 5q31, consists of 16 genes and three pseudogenes, all highly conserved and each one hosting a CpG island. We have developed a pyrosequencing assay that recognizes part of the CpG islands of 17 out of the 19 members of the cluster. Gene 1 and pseudogene 19P were excluded from the analysis, as their homology with the remaining PCDHB members is not sufficient to include them in the same assay as already previously noted.7
The success of this pyrosequencing assay depends on the amplification and complete, accurate reading of all the 17 fragments corresponding to each PCDHB member. To design a set of primers that could faithfully amplify the target DNA, the 17 sequences were aligned to find the most conserved regions suitable for primer design (Figure 1a and Supplementary Figure 1). As the alignment revealed multiple base differences among the 17 sequences, we introduced a degenerate base in the reverse PCR primer. To verify that the introduction of a degenerate base did not produce biases in the PCR amplification of the targets and in the subsequent pyrosequencing reaction, we sequenced the PCR products resulting from an annealing temperature gradient and found no differences in the level of methylation (data not shown).24 Differences between the gene sequences were taken into account also to design two of the three sequencing primers (Table 1) as described.25
Protocadherin B cluster (PCDHB) target sequence. (a) Annotated sequence of the amplified PCDHB genomic fragments (after bisulfite modification and in the hypothesis of complete methylation) considered in the pyrosequencing-based assay. The base differences between the 17 genes are reported above the sequence; the 20 CpG doublets are highlighted in light blue and their numbering is indicated above the sequence. The position of the pyrosequencing primers for amplification, sequencing and methylation specific quantitative PCR (MSqPCR) primers is reported below the sequence. (b) For every CpG doublet involved in mismatches to be corrected, highlighted in light blue, a short string of bases is shown. Position of the nucleotides utilized as reference peaks to calculate the precise methylation value of these CpG doublets is underlined.
To test the reproducibility of the PCDHB assay, we have calculated the s.d. between replicated samples in the same pyrosequencing run and between different runs (Supplementary Table 2). Two DNA samples (one hypo- and the other hypermethylated) subjected to this analysis showed essentially identical levels of methylation, demonstrating the optimal performance of the assay.
Sequencing and Interpretation of the Results
A large part of the amplified fragments was sequenced by using three primers to analyze a total of 20 CpG sites.
To take into account the mismatch at base 154, we modified the default dispensation order established by the Pyro Q-CpG software by dispensing the different base immediately before the prevalent one. This operation was not necessary for the other mismatches, as the changed base was identical to the previous or to the following one, and contextually sequenced with the same dispensed base (Table 1).
The result of the PCDHB pyrosequencing methylation assay consists of three pyrograms from which the mean value of methylation of each of the 20 CpG sites analyzed in the 17 sequences is obtained.
The Pyro Q-CpG software utilized to analyze the pyrosequencing results is tailored to obtain the quantitative methylation levels of the CpG sites in a sequenced fragment derived from a univocally determined genomic region. In the case of the PCDHB cluster, the multiple alignments revealed that some base changes occur within the CpG sites. In some cases (CpG 7, 10, 12 and 20), the different base is a G or an A instead of the C of the CpG doublet. As the software cannot consider other variable bases besides C (methylation) and T (unmethylation), these base changes would not be detected. In the other cases (CpG 1, 3, 5, 15 and 20), the C of the CpG doublet is replaced by a T, which is not modified by sodium bisulfite. The presence of this type of mismatch hampered the direct reading of the results by the analysis software, as this, not distinguishing between native T and T derived from the bisulfite conversion of unmethylated C, would alter the estimated percentage of methylation at these doublets.
To overcome all these problems, we derived correct methylation level from the raw pyrosequencing data, considering the peak height of the C of these CpG sites with respect to the height of a reference peak selected according to the following criteria: (i) it must be a unique base conserved in all 17 sequences; (ii) it can be any base, except A, whose emission is ‘adjusted’ by the instrument according to an internal algorithm and iii) it must be the nearest possible to the interrogated C. A value of 100% was imposed to the reference peak and the corrected percentage of methylation was calculated according to the following proportion: height of peak C: % methylation=height of reference peak: 100%. The eight CpG doublets that showed mismatches and the bases used as reference peaks utilized are indicated in Figure 1b.
Typical examples of pyrograms with the calculation of the percentage of methylation are reported in the Supplementary Figures 2a–f.
Our assay investigated 20 CpGs, 7 of which were originally considered for the definition of the PCDHB cluster as member of the methylator phenotype in NB by MSqPCR.7
To test the validity of our test, five samples, whose level of methylation was determined by pyrosequencing, were analyzed in parallel by MSqPCR and the calculated ‘methylation index’ was found to be comparable with the methylation value observed by pyrosequencing in the same CpGs (Figure 2).
For each patient, we compared by pyrosequencing the mean methylation level of the 20 CpG sites with that of the 7 CpGs originally considered by computing the Pearson's linear correlation coefficient. The perfect correlation between the two results indicates that the target sequence was homogeneously methylated (Figure 3a).
Correlation analysis between the methylation values of different regions of protocadherin B cluster (PCDHB) determined by pyrosequencing. (a) Correlation between the methylation values of 20 CpGs of the entire PCDHB fragment analyzed and the methylation value of the 7 CpGs considered in the original description of CpG island methylator phenotype (CIMP). (b–d) Correlation between the methylation values of the PCDHB fragments sequenced, respectively, with the S1 (b), S2 (c) and S3 (d) primers, and the methylation value of the seven CpGs considered in the original description of the CIMP.
Similar results were obtained by separately comparing the mean methylation level of the three sequenced regions with that of the seven CpGs from the MSqPCR analysis (Figures 3b–d).
Comparison of CIMP Methylation in INRG ‘Very Low-’ and ‘High- Risk’ Patients
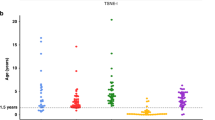
To verify if the pyrosequencing analysis of the PCDHB cluster could discriminate patients with different clinical characteristics, we have compared the methylation levels of a set of patients at stage 1, all alive and disease-free 5 years after the diagnosis, with those of patients at stage 4, who have died within 2 years from the diagnosis. According to the INRG criteria, these patients are classified into the very low- and high-risk group. Supplementary Table 1 shows the clinical characteristics of the patients included in the study and the mean PCDHB methylation values for each CpG site. In Figure 4 is shown the distribution of the methylation values of the PCDHB cluster in the two groups of patients. In the high-risk patients, the mean of the methylation was significantly higher than that of the very low-risk patients (Table 2). When the mean methylation value of the very low-risk patients (39,15%) was considered as a reference value, we observed that all patients of the high-risk group, except case 30, had a mean value of methylation higher than that of the lower risk group.
In an attempt to further simplify the test, we compared the two groups of patients considering separately the mean methylation values of the three sequenced regions, and statistically significant differences were reached in all cases (Table 2).
In particular, the results of mean methylation values derived from the region sequenced by primer S2 in very low-risk NB patients (40,10%) is comparable to the pyrosequencing results of the entire fragment (39.15%). Interestingly, these values, which represent the putative threshold discriminating the high-risk from the very low-risk NB patients in our study, are essentially identical to that resulting from the MSqPCR analysis,7 and are in full agreement with the relative Pearson's linear correlation coefficient of the same regions (Figure 3).
DISCUSSION
Hypermethylated DNA sequences are an extremely sensitive and specific tumor marker whose detection has been recently exploited as a system for the early diagnosis of primary cancer or of their recurrence.18, 19 The concordant hypermethylation of multiple genes in cancer is defined as CIMP and has been identified in many tumors including NB.4, 5, 6, 7, 8, 9, 10 In this tumor, the methylation of the PCDHB cluster, the most informative gene of the NB CIMP, is a promising biomarker of outcome and its predictive power was confirmed in independent sets of patients.7, 9 It is thus possible that if CIMP will be validated in prospective studies, it might be transferred to the clinical practice.
It has been recently demonstrated that the definition of threshold levels of methylation precisely identifies patients with different clinical characteristics.14 Indeed, for CIMP analysis, a quantitative method is required.5, 18, 19 In this respect, pyrosequencing is considered a gold standard technique for quantitative methylation studies, because the mean methylation of each CpG taken into consideration is expressed directly as an absolute value with great reproducibility, sensibility and accuracy.
The pyrosequencing assay that we have developed not only possesses these characteristics, but it can also simultaneously analyze the methylation level of 17 genes of the PCDHB cluster.
The great reproducibility and the direct sequencing of the pyrosequencing technology allow the precise quantification of DNA methylation and also disclose minimal differences of methylation levels. This latter feature is fundamental to identify the thresholds of methylation that, like in the case of PCDHB cluster in the NB, can be predictive of prognosis and help to classify the patients into distinct clinical groups.
A general potential drawback of the highly sensitive MSqPCR is that the accuracy of quantification is limited when evaluating narrow ranges of methylation.26 These limits have to be carefully taken into account for samples whose PCDHB methylation level is close to the threshold of methylation discriminating between patients at different risk.
The lack of standardization of the methylation assays makes difficult the comparison of the results among independent studies. Differences between methylation results in comparable groups of patients could result from the analysis of distinct regions within the CpG island. Therefore, before establishing a priori the target of a methylation study, a larger part of the sequence should be tested.27 By pyrosequencing, we have analyzed a longer DNA sequence in the PCDHB genes revealing that, in our experimental conditions, no polymorphisms in the sequences and no methylation hotspot that could alter the calculated methylation, were present. The ‘site-per-site’ information led us to establish that a smaller part of the sequence could be sufficiently informative to perform a simplified version of the test in the NB, an important finding in view of the clinical translation of our test. On the contrary, this preliminary elaboration is impossible with MSqPCR, because the result obtained is the mean value of methylation of only the CpG sites interrogated by the two couples of primers, independently from the methylation status of the sequence between the primers.
A general advantage of pyrosequencing is that a standard curve is unnecessary to calculate the mean of methylation in a target sequence. As a consequence, up to 96 samples can be sequenced in a single plate. A comparable number of samples processed in MSqPCR to study the PCDHB cluster would require more plates, because the two qPCR reactions, one for the methylated and one for the unmethylated allele, each with its technical replicates and its own standard curves,7, 28 use different annealing temperatures. Moreover, to calculate the ‘methylation index’ for a single sample by MSqPCR, it is necessary to set two triplicate reactions (three for the methylated and three for the unmethylated target); thus, even if the cost of the single pyrosequencing reaction is higher, the final cost of the two type of assays would be comparable. Moreover, the cost of pyrosequencing reaction and the amount of needed DNA can be further reduced, with respect to our protocol, depending on the type of pyrosequencing instrument and on the methylation analysis strategies utilized.29
The two standard curves necessary for a quantitative result in MSqPCR are an additional cost and contribute to limit the number of samples that can be processed at the same time.
Finally, the combination of two qPCR results coming from two different sets of primers could be an additional drawback of MSqPCR, because the reaction efficiency must be identical to combine the two sets of data to calculate the final ‘methylation index’.
The cost of the pyrosequencing instrument is the major limitation of our assay. However, because of the wide range of pyrosequencing-based diagnostic applications, this technology is becoming very common in clinical and research settings.
The standardization of an assay is an essential requirement for the transferability to the clinical practice. In this respect, the pyrosequencing technology is easy to perform and the procedure is partially automated, avoiding the variability due to the manual handling of the reagents.
Differently from MSqPCR technique, there are standard kits to perform the pyrosequencing reactions, an obvious advantage for the reproducibility of the results.
Pyrosequencing and MSqPCR are based on the chemical modification of DNA with sodium bisulfite to disclose the presence of the 5-methylcytosine in CpG doublets.
We have previously tested the reproducibility of pyrosequencing methylation analysis comparing the methylation levels of different genes in independent experiments and we observed a strong correlation (>95%) among the different repetitions.14 This reproducibility of the results is due not only to the robustness of the technique, but also to the internal controls that monitor the completeness of the chemical conversion with sodium bisulfite within the run. Because the chemical conversion of the DNA is not homogeneous in the genome and depends on the sequences characteristics, on the density of the CpG sites and on the methylation level, the built-in quality test of the chemical conversion of the target sequence avoids overestimating the methylation level in the case of incomplete conversion. This type of direct control is not present in MSqPCR.
Because of all these characteristics, we believe that pyrosequencing is an ideal tool to generate data that can be easily compared between different laboratories and data sets.
To determine if the assay could discriminate patients with different clinical characteristics and outcome, we have measured the level of methylation in NB patients at very low risk and at high risk.
The classification of the NB patients into the appropriate risk group is one of the criteria utilized to choose the optimal treatment regimen.20
Our results demonstrate that the levels of methylation of the PCDHB genes are significantly different between the two groups of NB patients at the opposite ends of the INRG20 classification system, and thus selected to be completely different in terms of outcome and disease progression.
Importantly, the methylation level that best discriminates between very low- and high-risk patients in our assay was essentially identical to that predicting poor survival in the original NB CIMP description, suggesting that the two assays are strongly, technically and biologically concordant.
In agreement with previous studies,7, 9 we retain the PCDHB cluster as a promising biomarker in NB, whose prognostic value should be precisely determined in multivariate analysis to take into account all variables known to influence the outcome of the NB patients. In this respect, the assay that we have developed might be an optimal option to achieve this result. PCDHB cluster is hosted in a chromosomal region at 5q31 and it is part of a larger PCDH@ cluster family, which also includes PCDHA and PCDHG clusters. These clusters are involved in the long-range epigenetic silencing, a coordinate event acting on large chromosomal regions30 and whose effects might resemble those of a chromosomal microdeletion.
In NB, epigenetic marks of repression are present not only in PCDHB, but also in the PCDHA cluster,7, 10 whereas no information is available for PCDHG. Interestingly, the PCDH@ family is aberrantly methylated in Wilms tumor and breast cancer;11, 12 thus, even if it has not been systematically examined, PCDH@ methylation appears as a novel epigenetic biomarker in different tumors. In this respect, the pyrosequencing-based assay that we have developed might be ideal not only for the fast and accurate analysis of the PCDHB cluster in NB, but also in other malignancies and, in principle, the strategy here delineated could be applied to other highly homologous gene clusters and particularly to the other members of the PCDH@ family in large patient sets.
References
Feinberg AP, Ohlsson R, Henikoff S . The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006;7:21–33.
Feinberg AP, Tycko B . The history of cancer epigenetics. Nat Rev Cancer 2004;4:143–153.
Jones PA, Baylin SB . The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415–428.
Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999;96:8681–8686.
Issa JP . CpG island methylator phenotype in cancer. Nat Rev Cancer 2004;4:988–993.
Teodoridis JM, Hardie C, Brown R . CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett 2008;268:177–186.
Abe M, Ohira M, Kaneda A, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res 2005;65:828–834.
Abe M, Watanabe N, McDonell N, et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology 2008;74:50–60.
Abe M, Westermann F, Nakagawara A, et al. Marked and independent prognostic significance of the CpG island methylator phenotype in neuroblastomas. Cancer Lett 2007;247:253–258.
Ushijima T, Okochi-Takada E . Aberrant methylations in cancer cells: where do they come from? Cancer Sci 2005;96:206–211.
Dallosso AR, Hancock AL, Szemes M, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms′ tumor. PLoS Genet 2009;5:e1000745.
Novak P, Jensen T, Oshiro MM, et al. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res 2008;68:8616–8625.
Alaminos M, Davalos V, Cheung NK, et al. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J Natl Cancer Inst 2004;96:1208–1219.
Banelli B, Bonassi S, Casciano I, et al. Outcome prediction and risk assessment by quantitative pyrosequencing methylation analysis of the SFN gene in advanced stage, high-risk, neuroblastic tumor patients. Int J Cancer 2010;126:656–668.
Banelli B, Gelvi I, Di Vinci A, et al. Distinct CpG methylation profiles characterize different clinical groups of neuroblastic tumors. Oncogene 2005;24:5619–5628.
Yagyu S, Gotoh T, Iehara T, et al. Circulating methylated-DCR2 gene in serum as an indicator of prognosis and therapeutic efficacy in patients with MYCN nonamplified neuroblastoma. Clin Cancer Res 2008;14:7011–7019.
Banelli B, Di Vinci A, Gelvi I, et al. DNA methylation in neuroblastic tumors. Cancer Lett 2005;228:37–41.
Kagan J, Srivastava S, Barker PE, et al. Towards clinical application of methylated DNA sequences as cancer biomarkers: A Joint NCI's EDRN and NIST Workshop on Standards, Methods, Assays, Reagents and Tools. Cancer Res 2007;67:4545–4549.
Duffy MJ, Napieralski R, Martens JW, et al. Methylated genes as new cancer biomarkers. Eur J Cancer 2009;45:335–346.
Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol 2009;27:289–297.
Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol 1993;11:1466–1477.
Tonini GP . Neuroblastoma: a multiple biological disease. Eur J Cancer 1993;29A:802–804.
Tost J, Gut IG . DNA methylation analysis by pyrosequencing. Nat Protoc 2007;2:2265–2275.
Shen L, Guo Y, Chen X, et al. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 2007;42:48, 50, 52 passim.
Dupont JM, Tost J, Jammes H, et al. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem 2004;333:119–127.
Oki Y, Aoki E, Issa JP . Decitabine—bedside to bench. Crit Rev Oncol Hematol 2007;61:140–152.
Nakagawachi T, Soejima H, Urano T, et al. Silencing effect of CpG island hypermethylation and histone modifications on O6-methylguanine-DNA methyltransferase (MGMT) gene expression in human cancer. Oncogene 2003;22:8835–8844.
Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32.
Paliwal A, Vaissiere T, Herceg Z . Quantitative detection of DNA methylation states in minute amounts of DNA from body fluids. Methods 2010;52:242–247.
Clark SJ . Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet 2007;16 (Spec No 1):R88–R95.
Acknowledgements
This work was supported by the Italian Ministry of Health, Core Grant to the Istituto Nazionale per la Ricerca sul Cancro (IST). BB is the recipient of the ‘Young Investigators’ Grant GR-2008-1143408 from the Italian Ministry of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
A novel pyrosequencing technique is described for methylation analysis of gene clusters with high internal homologies. The study confirms the predictive power of this technique for determining neuroblastoma methylator phenotype.
Supplementary information
Rights and permissions
About this article
Cite this article
Banelli, B., Brigati, C., Di Vinci, A. et al. A pyrosequencing assay for the quantitative methylation analysis of the PCDHB gene cluster, the major factor in neuroblastoma methylator phenotype. Lab Invest 92, 458–465 (2012). https://doi.org/10.1038/labinvest.2011.169
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2011.169
Keywords
This article is cited by
-
Combination of a synthetic retinoid and a DNA demethylating agent induced differentiation of neuroblastoma through retinoic acid signal reprogramming
British Journal of Cancer (2021)
-
Dysregulation of p53-RBM25-mediated circAMOTL1L biogenesis contributes to prostate cancer progression through the circAMOTL1L-miR-193a-5p-Pcdha pathway
Oncogene (2019)
-
DNA methylation profiling of primary neuroblastoma tumors using methyl-CpG-binding domain sequencing
Scientific Data (2016)
-
Clinical application of the CpG island methylator phenotype to prognostic diagnosis in neuroblastomas
Journal of Human Genetics (2013)