Abstract
Purpose:
Dysmorphology syndromes are among the most common referrals to clinical genetics specialists. Inability to match the dysmorphology pattern to a known syndrome can pose a major diagnostic challenge. With an aim to accelerate the establishment of new syndromes and their genetic etiology, we describe our experience with multiplex consanguineous families that appeared to represent novel autosomal recessive dysmorphology syndromes at the time of evaluation.
Methods:
Combined autozygome/exome analysis of multiplex consanguineous families with apparently novel dysmorphology syndromes.
Results:
Consistent with the apparent novelty of the phenotypes, our analysis revealed a strong candidate variant in genes that were novel at the time of the analysis in the majority of cases, and 10 of these genes are published here for the first time as novel candidates (CDK9, NEK9, ZNF668, TTC28, MBL2, CADPS, CACNA1H, HYAL2, CTU2, and C3ORF17). A significant minority of the phenotypes (6/31, 19%), however, were caused by genes known to cause Mendelian phenotypes, thus expanding the phenotypic spectrum of the diseases linked to these genes. The conspicuous inheritance pattern and the highly specific phenotypes appear to have contributed to the high yield (90%) of plausible molecular diagnoses in our study cohort.
Conclusion:
Reporting detailed clinical and genomic analysis of a large series of apparently novel dysmorphology syndromes will likely lead to a trend to accelerate the establishment of novel syndromes and their underlying genes through open exchange of data for the benefit of patients, their families, health-care providers, and the research community.
Genet Med 18 7, 686–695.
Similar content being viewed by others
Introduction
Although the scope of contemporary clinical genetics has greatly expanded beyond diagnosing rare dysmorphology syndromes, it remains an integral part of a typical clinical genetics practice. These syndromes often represent the pleiotropic effect of single gene mutations, and much has been learned about the function of human genes through the study of these syndromes despite their individual rarity. Pattern recognition is a key skill in the practice of clinical genetics because, although individual dysmorphic features can be part of the normal interindividual variation, it is their combined presence in an individual that prompts the diagnosis of a syndrome.1 Failure to find a match for the dysmorphology pattern in the published literature may not necessarily indicate the novelty of the syndrome because the matching process remains a largely subjective approach that is error-prone despite recent efforts toward standardization.2 In forums examining such cases (e.g., dysmorphology conferences and small meetings, databases, published literature), researchers often seek to establish that a particular syndrome is truly novel. Unfortunately, this process lacks throughput, and for many patients it is not unusual for many years to pass before they are designated as having a novel recognizable syndrome. Such designation is important, however, because it can be the basis for establishing the molecular pathogenesis and natural history of the disease through the identification of similarly affected patients.
Genotyping first is a recent trend made possible by the advent of genomic tools, initially in the form of genome-wide copy-number analysis and, more recently, sequencing tools that have the power to identify the likely causal mutation (genotype) regardless of knowledge of the disease (phenotype).3,4 Much has been published about the rapidly changing practice of clinical genetics as a consequence of next-generation sequencing technology, and about how the bottleneck has now shifted to identifying phenotypic matches for syndromes in which a novel candidate gene is identified based on a single family. This, in turn, will establish these as recognizable syndromes and facilitate reporting their candidate causal variants in the literature for the benefit of other patients, their caregivers, and the research community at large.5,6
Efforts have been made to accelerate the establishment of novel candidate genes in the literature. For example, we have published the identification of several novel candidate genes in the setting of retinal dystrophy and primordial dwarfism, and some of these genes have since been independently verified by others.7,8,9 Recently, we published the identification of 33 novel candidate genes for various neurocognitive phenotypes, and at least 6 of these were independently found to be mutated by other investigators in the few months since the article appeared online (unpublished data).10 However, we are not aware of any similar effort to accelerate the discovery of novel candidate genes specifically in the setting of dysmorphology syndromes. In this article, we describe our experience with members of 31 multiplex consanguineous families who appeared to have novel dysmorphology syndromes at the time of their initial evaluation; 15 of them are reported here for the first time. Genomic analysis of this cohort revealed novel disease candidates, and their reporting should facilitate “matchmaking” among the wider clinical genetics community.
Materials and Methods
Human subjects
Patients were evaluated as part of a standard clinical genetics evaluation by board-certified clinical geneticists. Eligible patients were those with an apparently novel phenotype involving, but not limited to, facial dysmorphism or skeletal dysplasia, positive family history consistent with autosomal recessive inheritance, and consanguineous parents. Two families (families 14 and 15) did not meet these criteria but were included because they have a clinical phenotype similar to that of family 13 (see below). Informed consent was obtained from all subjects prior to enrollment under an institutional review board–approved research protocol (KFSHRC RAC#2080006). Venous blood was collected in ethylenediaminetetraacetic acid for DNA extraction, and clinical photographs were taken after obtaining a separate photo consent form.
Autozygome analysis
Determination of the entire set of autozygous intervals per genome (autozygome) was as previously described.11 Briefly, we genotyped DNA samples using Axiom SNP Chip according to the manufacturer’s instructions (Affymetrix), followed by a genome-wide search for autozygous intervals using regions of homozygosity of >1 Mb as surrogates on AutoSNPa.12 When multiple affected members were available, their shared autozygome was determined.13
Whole-exome and whole-genome sequencing
Exome capture was performed using a TruSeq Exome Enrichment kit (Illumina, San Diego, CA) following the manufacturer’s protocol. Samples were prepared as an Illumina sequencing library; in the second step, the sequencing libraries were enriched for the desired target using the Illumina Exome Enrichment protocol. The captured libraries were sequenced using Illumina HiSeq 2000 Sequencer. The reads are mapped against UCSC hg19 (http://genome.ucsc.edu/) by Burrows-Wheeler Aligner (BWA) (http://bio-bwa.sourceforge.net/). The SNPs and Indels were detected by SAMTOOLS (http://samtools.sourceforge.net/). For whole-genome sequencing, amplification-free Illumina TrueSeq libraries were prepared, pooled, and then sequenced on six different Illumina HiSeq runs. Full-length paired-end reads were aligned using Burrows-Wheeler Aligner (BWA) MEM to Homo sapiens GRCh37 reference sequence (1000 Genomes Project phase 2: http://www.1000genomes.org/) with default parameters. BWA output was directly BAM-converted and genomic coordinate-sorted, and then subjected to a GATK insertion/deletion realignment process. We obtained average genome coverage of 13.11× (5×: 0.9590, 10×: 0.7338, 15×: 0.3022). Variants were called using both GATK’s UnifiedGenoTyper 3.2-2 and HaplotypeCaller 3.2-2. Both sets were filtered to remove variants that were present in more than 50% of individuals, those with less than three reads coverage, and those with more than 2.5% minor allele frequency in 1000 Genomes phase 3 release or the Exome Variant Server (http://evs.gs.washington.edu/EVS/). For both whole-exome and whole-genome sequencing, the candidacy of the resulting variants was based on their physical location within the autozygome of the affected individual, their population frequency, and the predicted effect on the protein as described previously.10,14
Results
Clinical characterization of apparently novel dysmorphology syndromes
Each of the 31 study families had a unique set of dysmorphic features and other systemic manifestations that did not appear to fit a previously recognized syndrome at the time of the analysis ( Table 1 and Figure 1 ; Supplementary Table S1 online). Family pedigrees for the 15 families reported here for the first time are shown in Figure 2 . In some families—e.g., 11DG0424, 14DG1221, and 11DG0268—there was a sufficient number of affected and unaffected members to map the phenotype to a single novel locus each, thus confirming the novelty of the phenotype even before exome sequencing. In most families, however, novelty of the phenotype could be verified only after exome sequencing.
Representative clinical images of the study subjects. (a) Clinical photograph for case 11DG0424 (family 1) showing microcephaly, coloboma, and prominent nasal bridge. (b) Clinical photograph for case 13DG0784 (family 2) showing very large ears, large nose, and deep-set eyes. (c) Clinical photograph for case 10DG1767 (family 3) showing a narrow chest with pectus carinatum. (d) Clinical photograph for case 10DG0648 (family 5) showing the deviated nasal septum and strabismus. (e) Clinical photograph for case 14DG1447 (family 7) showing retracted upper face, down-slanting palpebral fissures, and small nose. (f) Clinical photograph for case 13DG0916 (family 9) showing upturned nose and tented upper lip. (g) Clinical photograph for case 12DG1565 (family 10) showing unilateral left ptosis, hypoplastic maxilla, short upturned nose, and tented upper lip. (h) Clinical photograph for case 14DG1221(family 11) showing hypertelorism, strabismus, hypoplastic maxilla, micrognathia, low-set ears, and broad nose. (i) Clinical photograph for case 13DG1395 (family 12) showing severe microcephaly, hypertelorism, malar hypoplasia, low-set ears, wide nasal bridge, and bilateral cleft lip and palate. (j) Clinical photograph for case 14DG0993 (family 13) showing a stillborn boy with severely hypoplastic nose and abnormal genitalia. (k) Clinical photograph for case 15DG0764 (family 16) showing macrocephaly and bulbous nose. (l) Clinical photograph for case 10DG1175 (family 19) showing synophrys, deep-set eyes, bulbous nose, and prominent cheeks. (m) Clinical photograph for case 12DG0685 (family 21) showing hypoplastic maxilla and macrostomia with full lips. (n) Clinical photograph for case 11DG0502 (family 22) showing bilateral exophthalmos, strabismus, upturned nares, and long philtrum. (o) Clinical photograph for case 12DG1149 (family 23) showing severe ocular hypertelorism with mild synophrys and arched bushy eyebrows, infra-orbital creases, and depressed nasal bridge. (p) Clinical photograph for case 10DG1670 (family 29) showing full lips and prominent philtrum. (q) Clinical photograph for case 14DG0805 (family 17) showing severe microtia (upper limb deformity caused by absent radius is not visible). (r) Radiological image for case 08DG00384 (family 4) showing bilateral knee dislocation. (s) Radiological image of 13DG0792 (family 8) showing acromelia and metaphyseal and distal digital changes. (t) Clinical photograph of case 12DG1638 (family 6) showing severe syndactyly of the fingers.
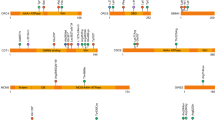
Pedigrees for the 15 families described for the first time in this article. The index is indicated in each pedigree by a black arrow. Asterisks denote individuals whose DNA was available for analysis and segregation. Blue boxes indicate the cases that were clinically evaluated. The hash tag (#) denotes individuals whose DNA was exome-sequenced.
High yield of autozygome/exome analysis for dysmorphology syndromes in multiplex consanguineous families
A strong candidate variant was identified in 90% (28/31) of the study cohort ( Tables 2 and 3 ). At a minimum, the causal variant must have been the only novel homozygous coding/splicing variant predicted to be pathogenic within the shared autozygome of the affected members of the respective family. In the remaining three families more than one variant remained, so we classified them as “unsolved.” Segregation of the variants was confirmed by Sanger sequencing among all available family members.
Consistent with the clinical impression that the phenotypes in this cohort are novel, only 19% (6/31) were found to have the strong candidate mutation in a known disease gene ( Table 3 ). In some families, this is because the previously published gene had an extremely limited phenotypic description. For example, family 21 (12DG0685) had a distinct constellation of features but their underlying disease gene ZNF526 was described only in the context of intellectual disability, with very few clinical details.15 In another family—family 24 (08DG00198)—the dysmorphology profile fit a recognizable but very rarely described syndrome, CDGIIa. Similarly, family 7 (14DG1447), which had a homozygous truncating mutation in MAN2B1, presented with nonspecific developmental delay and severe craniosynostosis. Craniosynostosis has very rarely been described in mannosidosis, so this diagnosis was not considered initially (Supplementary Figure S4 online). Finally, the phenotype for some genes was sufficiently different from what has been described in the literature that it was not possible to recognize them clinically. This includes family 3 (10DG1767), with members who presented with narrow chest and myopia and were later found to have cone–rod dysfunction. These members mapped to two autozygous intervals, neither of which appeared to contain a good candidate. Exome sequencing did not reveal any candidate coding/splicing variant. Close examination of the known disease genes within the two critical intervals highlighted C21orf2, an established disease gene for cone–rod dystrophy.7 Interestingly, one of the two C21orf2-linked cone–rod dystrophy families that we had originally described7 was found on careful examination to display short stature and narrow chest whereas the other was completely nonsyndromic. Thus, C21orf2 appears to cause both syndromic and nonsyndromic cone–rod dysfunction. We therefore carefully considered all novel homozygous variants in C21orf2 and identified a deep intronic mutation, which we confirmed as impairing normal splicing (Supplementary Figure S2 online). Similarly, family 27 (11DG0268) mapped to a single locus containing the known disease gene COG6, but the phenotype (intellectual disability and anhidrosis) was very different from the published COG6-related CDG phenotype, as we described in detail elsewhere.16,17
In 67% (21/31) of families, the strong candidate variant was identified in a gene that was novel at the time of analysis. These include 11 genes that we published previously10 and 10 that we describe for the first time here ( Table 2 ).
Family 1 (11DG0424 and 13DG2294) consists of two cousins with a strikingly similar phenotype that is best described as CHARGE-like. They both had coloboma, renal malformation, restricted growth, and limb anomalies ( Figures 1 and 2 ; Supplementary Table S1 online). A single autozygous interval was exclusively shared by the two affected corresponding to chr9:111,576,346-132,018,909, and therein the only novel candidate variant was a missense variant in CDK9 (NM_001261.3: c.673C>T; p.Arg225Cys, PolyPhen=probably damaging (0.913), Sorting Intolerant From Tolerant (SIFT)=deleterious (0.05)) (Supplementary Figure S1 online; Tables 1 and 2 ).
Family 2 (13DG0784) has a highly unusual dysmorphic syndrome characterized by very large ears, deep-set eyes, and severe developmental delay and growth deficiency ( Figure 1 ; Supplementary Figure S2 online). The homozygous truncating variant in ZNF668 (NM_001172668.1:c.955C>T; p.Gln319*) was the only novel candidate within the autozygome of the index ( Table 2 )
Family 4 (08DG00382 and 08DG00384) consists of two siblings with an apparently unique form of skeletal dysplasia with multiple joint dislocation ( Figure 1 ; Supplementary Figure S2 online). Exome sequencing in both siblings failed to identify a mutation in any of the genes known to cause skeletal dysplasia but revealed a novel missense variant in TTC28 (NM_001145418.1:c.1462G>A; p.Gly488Ser; PolyPhen=probably damaging (0.973); SIFT=deleterious (0)) as the only novel coding/splicing variant within the shared autozygome ( Table 2 ).
Family 5 (10DG0648, 10DG1459, and 10DG1460) consists of four affected siblings with progressive ataxia, developmental delay, and facial dysmorphism ( Figures 1 and 2 ; Supplementary Figure S3 and Table S1 online). Exome sequencing of one sibling did not reveal any novel variant within the shared autozygome between the three living affected siblings. Therefore, we proceeded with WGS, which revealed a homozygous microdeletion of 15,500 bp (hg19, chr13:96,442,001-96,457,500) that includes part of intron 11, exon 12, and the 3-untranslated region of DNAJC3 and part of the 3-untranslated region of UGTT2 (Supplementary Figure S3 online; Table 2 ). Synofzik et al.18 recently reported that DNAJC3 mutations cause progressive ataxia and neurodegeneration; therefore, this gene is not included among the 10 novel genes in this study.
Family 6 (12DG1638, 12DG2364, and 12DG2365) consists of a boy and his two sisters with severe syndactyly and variable penetrance of multiple pterygium ( Figures 1 and 2 ; Supplementary Figure S4 and Table S1 online). Three autozygous intervals (chr10:49,083,380-64,516,338, chr3:116,530,905-133,802,238 and chr11:95,069,943-105,483,626) were exclusively shared by the affected siblings. Exome sequencing revealed a large genomic deletion within the first locus that completely removes MBL2, which was further confirmed by high-resolution molecular karyotyping (Supplementary Figure S4 online). No other novel variants were identified by exome in either of the two remaining autozygous intervals.
Family 9 (13DG0916 and 15DG0234) consists of three siblings who presented with global developmental delay and variable penetrance of cleft palate and hypoglycemia ( Figure 1 ; Supplementary Figure S5 and Table S1 online). The splicing variant identified by exome sequencing in CADPS (NM_003716.3:c.442-1G>C) was the only novel coding/splicing variant within the shared autozygome of the two siblings who were alive. Reverse-transcription polymerase chain reaction (RT-PCR) using a patient-derived lymphoblastoid cell line confirmed that this homozygous mutation was truncating (p.Ile148Glnfs*24) (Supplementary Figure S5).
Family 10 (12DG1565 and 12DG1566) consists of two sisters with ptosis and facial dysmorphism ( Figure 1 ; Supplementary Figure S6 online). Exome sequencing revealed a homozygous truncating mutation in CACNA1H as the only novel coding/splicing variant within the shared autozygome ( Table 2 ).
Family 11 (14DG1221 and 15DG1187) consists of two affected members who presented with severe hypertelorism, high myopia, and, in one of the two, cleft lip and palate ( Figure 1 ; Supplementary Figure S6 and Table S1 online). A single autozygous interval (chr3:40899164-54379802) was exclusively shared by the two patients, in whom a missense variant in HYAL2 was identified (NM_033158.4:c.749C>T; p.Pro250Leu; PolyPhen=probably damaging (1); SIFT=deleterious (0)) (Supplementary Figure S6 online; Table 2 ).
Family 13 (14DG0993 and 15DG0395) consisted of three affected members with a unique syndrome characterized by facial dysmorphism, severe primary microcephaly with agenesis of corpus callosum, renal agenesis, congenital heart disease, and abnormal genitalia (Supplementary Figure S7 and Table S1 online). The index was exome-sequenced, and a homozygous synonymous variant (CTU2: NM_001012762.1:c.873G>A) was the only surviving variant that linked to the shared regions of homozygosity between two of the affected members. Subsequently, two additional families (families 14 and 15) with a nearly identical phenotype were recruited. The index from each of these two families was exome-sequenced and analyzed independently. Exome filtering revealed the same CTU2 variant as in family 13. Furthermore, the variant was linked to the only autozygous region shared between the affected members in the three families (chr16:88,155,503-89,588,896) with linkage LOD score of 4.5. RT-PCR revealed that the synonymous variant impairs the normal splicing with resulting frameshift and the introduction of a premature stop codon (NM_001012762.1: p.Thr247Alafs*21) (Supplementary Figure S8 online).
Family 16 (15DG0764 and 15DG0765) consisted of two affected members who presented with macrocephaly, hypoplastic maxilla, and skeletal dysplasia ( Figure 1 ; Supplementary Figure S9 online). Exome sequencing revealed a homozygous missense mutation in C3ORF17 (NM_015412.3:c.280C>T; p.Arg94Cys) as the only novel coding/splicing variant within the shared autozygome.
Family 18 (10DG0300, 10DG0301, and 10DG0538) was previously published based on an apparently novel phenotype consisting of joint contracture, limited upward gaze, and Legg–Calvé–Perthes disease.19 Exome sequencing revealed a novel missense variant in NEK9 (NM_033116.4:c.2042G>A; p.Arg681His, PolyPhen=probably damaging (0.999), SIFT=deleterious (0); Table 2 ).
Discussion
We have previously shown that the combined use of autozygome/exome analysis in the setting of multiplex consanguineous families has a diagnostic yield of 75% for neurocognitive phenotypes and 81% for retinal dystrophies.7,10 This prompted us to examine the yield of this approach for dysmorphology syndromes in the same setting, i.e., multiplex consanguineous families. Another reason for limiting our study to multiplex families is that the presence of more than one affected individual with the same apparently novel phenotype facilitates the recognition of the core phenotypic features of the syndrome, notwithstanding the known phenomenon of clinical variability. Using this approach, we show that the yield was also high at 91%.
Very recently, Bloss et al.20 published their experience with 17 families in which the proband appeared to have novel phenotypes. Subsequent whole-exome sequencing revealed a strong candidate mutation in 60%, including five novel candidate genes and four known disease genes. However, with the exception of one case with suspected Opitz G/BBB, none of the patients reported by Bloss et al. had a dysmorphology syndrome. In addition, several of the probands lacked family history, so nongenetic causes (at least in the Mendelian sense) could not be ruled out. Therefore, our study is distinct from the one by Bloss et al. in two main aspects. First, a likely mode of inheritance (autosomal recessive) was present in all of our study families. This may explain, at least in part, the higher yield of our study compared with that by Bloss et al. because this has also been shown in a recent review by the Centers of Mendelian Genomics.21 However, we note here that we cannot exclude the possibility of dual diagnosis and the presence of more than one underlying causal mutation in consanguineous pedigrees as described recently.22 Second, a distinct dysmorphology profile is present in each of the study families, and this will be much more likely to facilitate “matchmaking” as compared with some of the nonspecific phenotypes reported by Bloss et al. (e.g., developmental delay and muscle atrophy).
Despite our best effort to label a dysmorphology phenotype as novel only after an extensive literature search, we note that six study families were found to have mutations in known genes. In the case of MGAT2, we have previously shown that this is probably due to the poor documentation of dysmorphology in the metabolic literature.13,16,23,24 Similarly, we note that the craniosynostosis we observed in the setting of mild global developmental delay in the two siblings with MAN2B1 mutation was initially considered a novel phenotype for the same reason. Although craniosynostosis is not listed as a known feature of mannosidosis in OMIM or review articles, we found, in retrospect, that craniosynostosis had indeed been described, albeit very rarely.25,26
Our study describes 10 novel candidate genes, the candidacy of which is supported by multiple lines of evidence. First, the discovery of these candidates followed the same methodology we used for the other genes we identified in this study that were novel at the time of discovery but had since been confirmed by others (e.g., WWOX). Similarly, we have identified genes that had been independently identified by others (e.g., C11ORF46, CACNA1G, and ZNF526). Second, some of the novel candidates we report here are supported by compelling positional mapping data, e.g., CDK9, HYAL2, and CTU2 (each corresponding to a novel single critical locus on autozygosity mapping). Third, the candidacy of some of these genes is supported by available animal models. For example, Cadps is known to play an important role in regulating glycemic control in mouse.27 Similarly, Hyal2-deficient mice have craniofacial anomalies reminiscent of those observed in the two patients we describe.28 However, Cacna1h-deficient mice have no reported ptosis but rather cardiac fibrosis, which our two patients with CACNA1H homozygous truncation do not have.29 Interestingly, heterozygous missense changes in CACNA1H have been associated with childhood epilepsy, but the two sisters we describe with a homozygous truncating mutation in this gene are completely normal neurologically except for ptosis.30 This may suggest a different disease mechanism than we have previously shown for other genes in which biallelic loss of function results in distinct clinical phenotypes as compared with heterozygous mutations.9,31,32 Alternatively, the susceptibility to absence seizures may have been erroneous.
The relatively easy access to genomic sequencing tools has empowered many clinical geneticists to identify interesting novel candidate genes in their patients. However, many of these tentative links have not been published because they are typically retained until a second case with a matching phenotype and genotype is identified. Several matchmaking tools have been developed to address this bottleneck, e.g., GeneMatcher and MIMmatcher.33,34 Reporting detailed clinical and genomic analyses of a large series of apparently novel dysmorphology syndromes will likely lead to a trend of accelerating the establishment of novel syndromes and their underlying genes. Such a trend will catalyze matchmaking such that the proposed novel phenotype is established and its candidate gene is confirmed independently. It is only through high-throughput identification and confirmation of disease–gene links that we can reap the benefits of full medical annotation of the human genome.
Disclosure
The authors declare no conflict of interest.
References
Aase JM. Diagnostic Dysmorphology. Springer: New York, 1990.
Carey JC, Allanson JE, Hennekam RC, Biesecker LG. Standard terminology for phenotypic variations: the elements of morphology project, its current progress, and future directions. Hum Mutat 2012;33:781–786.
Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell 2014;156:872–877.
Lupski JR. Structural variation mutagenesis of the human genome: Impact on disease and evolution. Environ Mol Mutagen 2015;56:419–436.
Desai AN, Jere A. Next-generation sequencing: ready for the clinics? Clin Genet 2012;81:503–510.
Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet 2013;14:681–691.
Abu-Safieh L, Alrashed M, Anazi S, et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res 2013;23:236–247.
Aldahmesh MA, Khan AO, Mohamed JY, et al. Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet Med 2012;14:955–962.
Shaheen R, Faqeih E, Ansari S, et al. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res 2014;24:291–299.
Alazami AM, Patel N, Shamseldin HE, et al. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep 2015;10:148–161.
Alkuraya FS. Autozygome decoded. Genet Med 2010;12:765–771.
Carr IM, Flintoff KJ, Taylor GR, Markham AF, Bonthron DT. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum Mutat 2006;27:1041–1046.
Alkuraya FS. Discovery of rare homozygous mutations from studies of consanguineous pedigrees. Curr Protoc Hum Genet 2012:Unit 6.12.
Alkuraya FS. The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Hum Genet 2013;132:1197–1211.
Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011;478:57–63.
Shaheen R, Ansari S, Alshammari MJ, et al. A novel syndrome of hypohidrosis and intellectual disability is linked to COG6 deficiency. J Med Genet 2013;50:431–436.
Alkuraya FS, Shaheen R. Variable phenotypic expression of COG6 mutations. J Med Genet 2014;51:425–426.
Synofzik M, Haack TB, Kopajtich R, et al. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet 2014;95:689–697.
Alkuraya FS. Arthrogryposis, perthes disease, and upward gaze palsy: a novel autosomal recessive syndromic form of arthrogryposis. Am J Med Genet A 2011;155A:297–300.
Bloss CS, Zeeland AA, Topol SE, et al. A genome sequencing program for novel undiagnosed diseases. Genet Med; e-pub ahead of print 19 March 2015.
Chong JX, Buckingham KJ, Jhangiani SN, et al.; Centers for Mendelian Genomics. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet 2015;97:199–215.
Yavarna T, Al-Dewik N, Al-Mureikhi M, et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum Genet 2015;134:967–980.
Alkuraya FS. Mental retardation, growth retardation, unusual nose, and open mouth: an autosomal recessive entity. Am J Med Genet A 2010;152A:2160–2163.
Alazami AM, Monies D, Meyer BF, et al. Congenital disorder of glycosylation IIa: the trouble with diagnosing a dysmorphic inborn error of metabolism. Am J Med Genet A 2012;158A:245–246.
Courtney RJ, Pennesi ME. Retinal dystrophy in 2 brothers with α-mannosidosis. Arch Ophthalmol 2011;129:799–802.
Grabb PA, Albright AL, Zitelli BJ. Multiple suture synostosis, macrocephaly, and intracranial hypertension in a child with alpha-D-mannosidase deficiency. Case report. J Neurosurg 1995;82:647–649.
Speidel D, Salehi A, Obermueller S, et al. CAPS1 and CAPS2 regulate stability and recruitment of insulin granules in mouse pancreatic beta cells. Cell Metab 2008;7:57–67.
Jadin L, Wu X, Ding H, et al. Skeletal and hematological anomalies in HYAL2-deficient mice: a second type of mucopolysaccharidosis IX? FASEB J 2008;22:4316–4326.
Chen CC, Lamping KG, Nuno DW, et al. Abnormal coronary function in mice deficient in alpha1H T-type Ca2+ channels. Science 2003;302:1416–1418.
Heron SE, Phillips HA, Mulley JC, et al. Genetic variation of CACNA1H in idiopathic generalized epilepsy. Ann Neurol 2004;55:595–596.
Aldahmesh MA, Mohamed JY, Alkuraya HS, et al. Recessive mutations in ELOVL4 cause ichthyosis, intellectual disability, and spastic quadriplegia. Am J Hum Genet 2011;89:745–750.
Patel N, Faqeih E, Anazi S, et al. A novel APC mutation defines a second locus for Cenani–Lenz syndrome. J Med Genet 2015;52:317–321.
Gottlieb MM, Arenillas DJ, Maithripala S, et al. GeneYenta: a phenotype-based rare disease case matching tool based on online dating algorithms for the acceleration of exome interpretation. Hum Mutat 2015;36:432–438.
Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat 2015;36:425–431.
Acknowledgements
We thank the patients and their families for their enthusiastic participation. We also thank the Genotyping and Sequencing Core Facilities at KFSHRC for their technical help. This work was supported in part by a King Salman Center for Disability Research grant (F.S.A.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
(PDF 6723 kb)
Supplementary Table S1
(DOC 512 kb)
Rights and permissions
About this article
Cite this article
Shaheen, R., Patel, N., Shamseldin, H. et al. Accelerating matchmaking of novel dysmorphology syndromes through clinical and genomic characterization of a large cohort. Genet Med 18, 686–695 (2016). https://doi.org/10.1038/gim.2015.147
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2015.147
Keywords
This article is cited by
-
Insight into ALKBH8-related intellectual developmental disability based on the first pathogenic missense variant
Human Genetics (2022)
-
Prenatal exome sequencing and chromosomal microarray analysis in fetal structural anomalies in a highly consanguineous population reveals a propensity of ciliopathy genes causing multisystem phenotypes
Human Genetics (2022)
-
Biallelic CDK9 variants as a cause of a new multiple-malformation syndrome with retinal dystrophy mimicking the CHARGE syndrome
Journal of Human Genetics (2021)
-
ZNF668 deficiency causes a recognizable disorder of DNA damage repair
Human Genetics (2021)
-
Genetics of intellectual disability in consanguineous families
Molecular Psychiatry (2019)