Abstract
Purpose: Keratoconus is a corneal dystrophy with an incidence of 1 in 2000 and a leading cause for cornea transplantation in Western developed countries. Both clinical observations and segregation analyses suggest a major role for genes in its pathogenesis. It is genetically heterogenous, most commonly sporadic, but inherited patterns with recessive or dominant modes have also been reported. We studied a four-generation autosomal-dominant pedigree to identify disease loci for keratoconus.
Methods: A two-stage genome-wide scan was applied to 27 family members. First linkage analysis was performed with 343 microsatellite markers along the 22 autosomal chromosomes at ≈10 cM density. This was followed by fine mapping at ≈2 cM density, in regions suggestive of linkage. Multipoint linkage analysis was performed using GeneHunter2.
Results: Evidence of suggestive linkage from the initial scan was observed at the 82 to 112 cM region of chromosome 5q14.1-q21.3 with a maximum lod score (LOD) of 3.48 (penetrance = 0.5). Fine mapping by testing an additional 11 microsatellite markers at 1 to 3 cM intervals revealed a narrower and higher peak (99–119 cM) with LOD 3.53. By analysis of the recombination of haplotypes, the putative locus of keratoconus was further narrowed to a 6 cM region (8.2 Mbp physical distance) between markers D5S2499 and D5S495.
Conclusion: These results indicate a promising new locus for keratoconus in this pedigree. Because of the heterogeneous nature of keratoconus, this locus may be specific to familial autosomal-dominant keratoconus. Nevertheless, the identification of this locus may provide new insights into the pathogenesis of keratoconus.
Similar content being viewed by others
Main
Keratoconus [MIM 148300] is a noninflammatory progressive corneal thinning disorder resulting in mixed myopia and irregular astigmatism. The progression of keratoconus may result in severe visual impairment and some affected individuals require cornea transplantation.1,2 The prevalence of keratoconus is about 1 in 2000 in Caucasian populations.1 It is a leading cause for cornea transplantation in Western developed countries.
Symptoms of keratoconus usually develop in the teens and are likely to progress during the early twenties, whereas progression slows after age 30.1,2 Keratoconus can be diagnosed by well-recognized clinical signs, including stromal thinning, Vogt's striae, Fleischer's ring, and scissoring of the retinoscopic reflex with a fully dilated pupil.3,4 The most sensitive and accurate diagnostic method is the computer-assisted videokeratography generated by devices such as the Topographic Modeling System (TMS-1). The TMS-1generates thousands of data points from the center of the cornea to the periphery for visual display. This system also automatically calculates multiple indices derived from data points in the central and paracentral cornea in one sitting. It can be used to detect keratoconus at an early stage before clinical signs are apparent.3,4
Although the etiology of keratoconus is unknown, several lines of evidence support an underlying genetic basis for this disease.5–8 This evidence includes the following: reports of keratoconus associated with other genetic disorders, such as Down and Marfan syndrome,9 observations of multigeneration pedigrees with either a dominant or recessive inheritance patterns with incomplete penetrance,1,7 and a reported 6% to 23.5% positive family history among patients with keratoconus.5,6 Furthermore, the estimated keratoconus prevalence in the first-degree relatives is 3.34%, 15 to 67 times higher than in the general population.5,7,10 Lastly, in the limited twin studies, 54% concordance rate among monozygotic twins was observed.11,12
In the majority of keratoconus patients (in excess of 90%), the disorder is bilateral. In many instances, the disorder may start of a unilateral in nature, but over time, the other eye becomes affected.8,13 Moreover, there are multiple case reports of recurrent keratoconus after cornea transplant.14–16 Such bilaterality and recurrent keratoconus occurring in cornea grafts also lends support to a genetic basis for this disease. However, the nature of genetic contribution to the development of keratoconus is complex. Genetic heterogeneity is suggested by the fact that both dominant and recessive models have been observed in keratoconus pedigrees5,17 and the majority of familial cases do not fit simple Mendelian inheritance patterns.
The genetic heterogeneity and the phenotypic diversity of keratoconus present challenges to the dissection of genetic factors in the development of keratoconus. Various approaches for identification of disease susceptibility genes have been applied to keratoconus and revealed several putative loci. These include a putative locus on chromosome 16q, identified in 20 Finnish small pedigrees by a genome-wide nonparametric linkage analysis18; a locus on 20q12, detected by a genome-wide identity-by-descent approach in a genetically isolated population of Tasmania, Australia19; a locus on chromosome 15q22.23-q24.2, identified in a large Northern Irish family20 and a locus on chromosome 3p14-q13, identified in an Italian two-generation autosomal-dominant pedigree by a genome-wide parametric linkage analysis.21 In addition, a candidate gene study in two Canadian families demonstrated the involvement of the VSX1 homeobox gene in keratoconus.22 These studies provide further support for a genetic basis for keratoconus and demonstrate the genetically complex nature of keratoconus. The aim of this study was to identify keratoconus loci in a four-generation Caucasian autosomal-dominant pedigree, using a two-stage, genome-wide scan approach. Our results demonstrate evidence for a novel locus for keratoconus at 5q14.3-q21.1.
MATERIALS AND METHODS
Study subjects and clinical evaluation
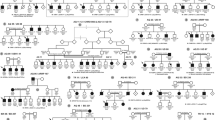
Twenty-seven subjects were recruited from a four-generation family with dominantly inherited keratoconus (Fig. 1). This family has previously been described in part and is currently being followed longitudinally under the protocol of the Keratoconus Genetics Research Program at Cedars-Sinai Medical Center.8,23 This research protocol was reviewed and approved by the Institutional Review Board of the Cedars-Sinai Medical Center. Informed consent was obtained from all study subjects.
Pedigree of a four-generation Caucasian family with haplotypes of seven markers. Generation number is indicated by the bold-faced Roman numerals listed to the left of the generation. Individual identification number of each member is indicated at the right bottom side of each member. Markers and their corresponding locations are indicated in the lower-left corner. Italicized numbers in haplotypes (e.g., IV-1 and IV-2) were deduced alleles from the parent's haplotype, and “−”denotes that the genotypes for those markers were not available.
All family members underwent clinical evaluation, which included slit lamp biomicroscopy, dilated retinoscopy, and computer-assisted videokeratography. Each subject additionally had a full ocular and systemic evaluation by a trained ophthalmic geneticist to rule out any known syndromes or other reported associations with keratoconus.24,25 Patients were assigned as having keratoconus if they had at least two of the following previously reported clinical signs of keratoconus: stromal corneal thinning (centrally or paracentrally), Fleischer's ring, Vogt's striae, scissoring, or an oil droplet sign on dilated retinoscopy. Patients were also assigned as having “subclinical” keratoconus, if they had a videokeratography pattern designated as an asymmetric bowtie with skewed radial axes (AB/SRAX) (Fig. 2), without the aforementioned clinical signs.24,25
Videokeratography methods and analysis
Videokeratography was performed on both eyes of each study subject with the TMS-1 (Computed Anatomy, Inc, New York, NY), software version 1.61. This software embodies an algorithm that uses axial radius of curvature calculations to display topographic maps. The following measures were taken to ensure quality control and reproducibility of video images: a minimum of four pictures of each eye were taken; the inner 22 of the 25 rings had to be fully digitized; lids had to be kept wide open and off the cornea without assistance or eyelid compression; all pictures had to be within 0.25 D of each other as measured by simulated keratometry (Sim K) readings; and to ensure proper fixation and alignment, the crosshair had to be in the center of the pupil for each examination. The best videokeratograph of the four was selected based on the quality of the keratoscope mires by visual inspection. Three members of our research team, who had no knowledge of the clinical status of the patient, then independently inspected each selected videokeratograph and assigned a pattern according to a videokeratograph scheme developed by our group26,27 (Fig. 2). This classification scheme has previously been demonstrated to be highly reliable and reproducible even among untrained observers.28 Agreement was necessary among the three observers to assign a videokeratograph a specific pattern. Videokeratographs were assigned as either having an AB/SRAX pattern (thus as affected) or one of the other patterns that form part of this classification scheme (designated as normal)25 (Table 1).
Genotyping
DNA was extracted from either blood samples or EBV-transformed lymphoblastoid cell lines following a standard protocol from Puregene Gentra Systems, Inc. (Minneapolis, MN).
A two-stage whole genome scan approach was used to identify keratoconus loci. For the initial stage, a total of 343 microsatellite markers from the ABI (Foster City, CA) 10 cM linkage panel, LMS-MD10, spanning the 22 autosomes at ≈10 cM density, was genotyped for all subjects. The heterozygosity of these markers ranged from 0.58 to 0.91, with an average of 0.79.
Markers were amplified using standard polymerase chain reaction (PCR) protocols with AmpliTaq Gold PCR Master Mix (ABI, Foster City, CA), appropriate fluorescent-labeled primer pairs, and 10 to 15 ng DNA in a GeneAmp PCR System 9700, (ABI, Foster City, CA). PCRs were multiplexed when different primer sets had common annealing temperatures and their product sizes varied significantly or they were labeled with different fluorescent dyes. The PCR products were separated on a 6% polyacrylamide gel, denatured with 6M urea, in an ABI Genetic Analyzer 377, and analyzed using the software Genotyper v2.5 (ABI, Foster City, CA).
After the initial scan under the most conservative scenario (D, see later), one region with suggestive linkage (LOD > 2) on chromosome 5q14.1-q21.3 was further fine mapped with markers at 1 to 2 cM intervals (Table 2). Fine mapping markers were selected from the databases of the Marshfield Human Genetic Maps (http://www.marshfieldclinic.org) and the Human Genome Resources of the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov). Eleven additional markers were typed for the 5q14.1-q21.3 region (approximate 30 cM), including D5S672, D5S641, D5S401, D5S618, D5S815, D5S2499, D5S2100, D5S484, D5S2079, D5S495, and D5S409 with heterozygosities ranging from 0.63 to 0.82 with an average of 0.74 (Table 2). The information content in the region ranged from 0.66 to 0.93 with an average of 0.88. The PCR primer pairs for these markers were designed using the software Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3_www.cgi) and ordered from Qiagen (Valencia, CA). The PCR products were analyzed in an ABI Genetic Analyzer3100 (ABI, Foster City, CA).
Statistical analysis
Our pedigree contains two families, connected by a mating between two affected individuals (III-4 and III-5, see Fig. 1). According to the knowledge of all participants in this study, the two families are not descendants from a common ancestry. The offspring of these two subjects may inherit the disease gene from either affected parent, and the genes from both parents may or may not be located in the same region. There is no prior knowledge or optimal strategy of how to use such a complex pedigree effectively. Thus, we conducted data analysis in several pedigree scenarios by dividing bilateral families at different individuals. These were as follows: (A) no division, which is an unbiased approach; (B) dividing the pedigree at III-5 so that children of III-4 and III-5 were grouped with maternal side; (C) dividing the pedigree at III-4 so that children of III-4 and III-5 were grouped with paternal side; and (D) drop all children of III-4 and III-5, which was the most conservative approach, but resulted in a great loss of significant information (Table 3).
Model-based multipoint linkage analysis was performed using the Markov Chain Monte Carlo (MCMC) method as implemented in SimWalk2 v2.8329 (for scenario A and C) and the Lander-Green method as implemented in GeneHunter v2.030(for scenario B and D). An autosomal-dominant model was assumed with a disease frequency = 1/2000 and various penetrance and phenocopy parameters. Under the conservative pedigree scenario (D), the region with LOD score > 2 was followed with fine mapping. The size of the region to be fine mapped was determined by covering the entire region with boundaries at LOD = 1.
The maximum LOD score was obtained by performing multipoint linkage analyses under four family structure scenarios and different parameters for penetrance (0.5 ≈ 0.9) and phenocopy (0.01, 0.05). The largest LOD score was observed under scenario B, penetrance = 0.5, and phenocopy = 0.01. In order to evaluate the statistical significance of this maximum LOD score, we calculated the empirical P values by simulating 500 replicates under the assumption of no linkage using SIMULATE.31 We simulated 500 replicates under scenario A (no division) for 15 markers in the linkage region with frequencies and map distance identical to the original data as shown in Table 2. Each of these replicates was divided into scenario B, C, and D as defined above. The highest LOD score for each replicate (Max-LOD) was obtained from linkage analyses performed for each replicate under all scenarios and combinations of parameters (phenocopy 0.01, 0.05, and penetrance 0.5–0.9). The empirical P values were computed by dividing the number of replicates that exceeded a particular Max-LOD threshold by the total number of replicates (500).
To determine the keratoconus risk haplotype, haplotypes in the linked region for all family members were constructed by using GeneHunter v2.0.30 The shared region of the haplotypes among all affected individuals may imply the location of a keratoconus locus. This analysis further narrowed a putative region linked with keratoconus.
RESULTS
Phenotype
Diagnostic information on all study subjects is summarized in Table 1. Fourteen individuals of the 27 studied family members were diagnosed with keratoconus by either clinical signs or subclinical videokeratographic pattern. Examples of the keratoconus subclinical diagnosis are II-4 and III-14, where both indicate a clear keratoconus VK pattern (AB/SRAX) (see Fig. 3), but did not have sufficient clinical signs of keratoconus. This four-generation pedigree is consistent with an autosomal-dominant inheritance pattern with reduced penetrance.
Initial genome scan and fine mapping
With an autosomal-dominant model, penetrance = 0.8, phenocopy = 0.01, we performed a multipoint linkage analysis using GeneHunter v.2.0 under the conservative scenario D. The region achieved the suggestive linkage (LOD > 2) is on chromosome 5q14.1-q21.3 with LOD = 2.10 (Fig. 4). After fine mapping of this region (≈30 cM) with markers at 1 to 2 cM density, the LOD score of the region on chromosome 5q14.1-q21.3 was increased to 2.18 under scenario D.
Maximum LOD score
Table 3 summarizes LOD scores under each of the four scenarios for both initial scan and fine mapping linkage analyses at chromosome 5q14.1-q21.3. Under both unbiased (A) and most conservative (D) scenarios, the evidence for suggestive linkage (LOD > 2) was obtained. Furthermore, under scenario C, where the pedigree was divided at III-4 so that affected children were grouped with the paternal family, LOD score during fine mapping was 0.88, whereas under scenario B, where the pedigree was divided at III-5 so that affected children were grouped with the maternal family, a maximum LOD score of 3.29 was obtained. Such results indicate that the affected children, IV-4, IV-5, and IV-6, are more likely to inherit the disease gene from their maternal ancestor rather than their paternal ancestor.
Using pedigree scenario B, we performed multipoint linkage analysis with several penetrances (0.5 to 0.9) and phenocopy parameters (0.01 and 0.05) for both initial scan and fine mapping markers on chromosome 5. Table 4 shows the maximum LOD scores and positions on chromosome 5 under different parameters by the GeneHunter program. Significant linkage was observed on chromosome 5q14.1-q21.3 with a maximum LOD score of 3.48 near marker D5S644 (82–112 cM) at penetrance = 0.5 and phenocopy = 0.01. After fine mapping the LOD score increased to 3.53 near marker D5S484 (Fig. 5B and Table 4). This peak was located between markers D5S401 and D5S2027, around 20 cM region. Fine mapping narrowed the range of the disease locus from 30 to 20 cM at 5q14.3-q21.3. The empirical P value for MAX-LOD > 2 is 0.006 (3/500). The empirical P value for MAX-LOD > 3 is < 0.002 because no MAX-LOD exceed 3.
Linkage results of chromosome 5 (penetrance = 0.5, phenocopy = 0.01) under pedigree scenario B. Each short line on the horizontal axis represents a marker corresponding to their locations. A, Initial scan, at ≈10 cM density. B, Fine mapping at 1 to 2 cM density in the region between D5S424 and D5S433. Name and position of each marker can be referenced the marker order on Table 2. LOD scores of each marker are plotted on the vertical axis.
Haplotype analysis
Haplotypes of each member were constructed for 11 markers in the linkage region (D5S401–D5S433), but 7 markers were adequate to illustrate the recombination events in the region as shown in Figure 1. In each of the two families, one haplotype cosegregated with keratoconus as illustrated in Figure 1 by the black or gray shading. That is, all affected individuals carry at least one disease haplotype. Some of the affected children from two affected parents III-4 and III-5 have both disease haplotypes. Recombination events were observed in individuals III-2, III-5, III-7, and III-9 and defined the critical keratoconus gene location as between markers D5S2499 and D5S495 (Fig. 6). These results indicate a novel locus for keratoconus in 5q14.3-q21.1, an approximate 8.2-Mbp region between markers D5S2499 and D5S495 for this pedigree.
DISCUSSION
In a four-generation bilineal keratoconus pedigree with an autosomal-dominant inheritance pattern, we identified a putative locus for keratoconus on chromosome 5q14.3-q21.1 between markers D5S2499 and D5S495. All affected individuals from each family share a common haplotype within each family (Fig. 1). The complexity of linkage analysis in this pedigree is its bi-lineage, so the power to identify linkage is reduced because the children of the two affected parents can inherit a disease gene from either parent. Under the most conservative scenario of dropping all children from the two affected parents, we observed a suggestive linkage on 5q14.3-q21.1 (LOD = 2.18), whereas the largest LOD score was observed under scenario B when dividing the pedigree at III-5 and the children were grouped with maternal family and penetrance was set as 0.5 (LOD = 3.53) (Table 3). To evaluate empirically the probability for observing this LOD score, we conducted simulation by optimizing over family structures and parameters (penetrance and phenocopy) and observed that the probability of identifying a MAX-LOD > 2 is 0.006, for a LOD > 3, it is < 0.002. Thus, the chromosome 5q14.3-q21.1 region is highly likely to contain a locus for keratoconus in this autosomal-dominant pedigree.
In our pedigree, we did not observe evidence for suggestive linkage (LOD > 2) for previously reported loci, including chromosome 16q22.3–23,18 20q12,19 3p14,21 and 20p1122 for keratoconus, and 15q22 for familial keratoconus with cataracts.20 Moreover, we could exclude two loci in this pedigree (LOD < −2), 3p14 and 16q22.3–23.
This four generation family with keratoconus demonstrates an autosomal mode of inheritance with variable expression and some interesting phenotypic features, similar to other autosomal dominant pedigrees. Clinically, all patients had “nipple type” or central cones of the cornea in various degrees of severity. For example, the disease was so severe in IV-4, IV-5, and IV-6 that they required corneal transplants in their early thirties. In contrast, for patients II-4 (Age 92) and III-14 (Age 42) their disease was so mild there were no clinical signs of the disease and nipple type cones were detected by videokeratography only. Both patients (II-4, III-14) had an AB/SRAX pattern that has been demonstrated in several studies to be an excellent and highly accurate videokeratograph recognition pattern for keratoconus.7,26,28
The significant linkage evidence (LOD = 3.53) and the fact that all affected individuals in this pedigree inherited the identical haplotype within each of the two families argue that there is likely to be a novel locus on 5q14.3-q21.1, which may be specific to this pedigree or a subset of familial dominant keratoconus. Considering all previously reported and this current linkage evidence in aggregate, it is clear that we are dealing with a genetically heterogeneous disorder although some of these loci may be the result of the type 1 error.
Although every affected individual in this pedigree carries at least one disease haplotype, there are some members (III-10, III-12, IV-3, IV-7, and IV-9), who carry the presumed disease haplotype but were not diagnosed with keratoconus (Fig. 1, Table 1). Such findings may be explained by reduced penetrance or indicate the need of more sensitive clinical tools in identifying subclinical abnormalities. The observation that the evidence for linkage reached the peak when penetrance was set at 0.5 indicates that the penetrance for this putative locus is modest.
Because keratoconus is a genetically heterogeneous disease, studying a single large family or an isolated population may reduce genetic heterogeneity and is more likely to be successful in revealing linkage as demonstrated here and in other reports.19,21 Any genes identified in large families or isolated populations may provide new insights into the pathogenesis of keratoconus in general. Within the 8.2-Mbp 5q14.3-q21.1 region linked in the current family there are approximately 50 known or predicted genes, which are currently being evaluated, including calpastatin (CAST), type 1 tumor necrosis receptor shedding aminopeptidase regulator (ARTS-1), and proprotein convertase subtilisin/kexin type 1 (PCSK1).
Genome-wide scan linkage analysis revealed a promising new locus for keratoconus in this multigeneration dominant keratoconus pedigree. Because of the heterogeneous nature of keratoconus, this locus may be specific to familial autosomal-dominant keratoconus. Nevertheless, the identification of this locus may provide new insights into the pathogenesis of keratoconus in general.
References
Rabinowitz YS . Keratoconus. Surv Ophthalmol 42: 297–319. 1998.
Gasset A, Hinson WA, Frias JL . Keratoconus and atopic diseases. Ann Ophthalmol 1978; 10: 991–994.
Wilson SE, Verity SM, Conger DL . Accuracy and reproducibility of the corneal analysis system and topographic modeling system. Cornea 1992; 11: 28–35.
Bawazeer AM, Hodge WG, Lorimer B . Atopy and keratoconus: a multivariate analysis. Br J Ophthalmol 2000; 84: 834–836.
Wang Y, Rabinowitz YS, Rotter JI, Yang H . Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet 2000; 93: 403–409.
Edwards M, McGhee CN, Dean S . The genetics of keratoconus. Clin Experiment Ophthalmol 2001; 29: 345–351.
Rabinowitz YS . The genetics of keratoconus. Ophthalmol Clin North Am 2003; 16: 607–620.
Rabinowitz YS, Maumenee IH, Lundergan MK, Puffenberger E, Zhu D, Antonarakis S et al. Molecular genetic analysis in autosomal dominant keratoconus. Cornea 1992; 11: 302–308.
Parker J, Ko WW, Pavlopoulos G, Wolfe PJ, Rabinowitz YS, Feldman ST . Videokeratography of keratoconus in monozygotic twins. J Refract Surg 1996; 12: 180–183.
Forstot SL, Goldstein JH, Damiano RE, Dukes DK . Familial keratoconus. Am J Ophthalmol 1988; 105: 92–93.
Bechara SJ, Waring GO III, Insler MS . Keratoconus in two pairs of identical twins. Cornea 1996; 15: 90–93.
Zadnik K, Mannis MJ, Johnson CA . An analysis of contrast sensitivity in identical twins with keratoconus. Cornea 1984; 3: 99–103.
Holland DR, Maeda N, Hannush SB, Riveroll LH, Green MT, Klyce SD et al. Unilateral Keratoconus incidence and quantitative topographic analysis. Ophthamology 1997; 140: 1409–1413.
Thalasselis A, Etchepareborda J . Recurrent keratoconus 40 years after keratoplasty. Ophthalmic Physiol Opt 2002; 22: 330–332.
Kremer I, Eagle RC, Rapuano CJ, Laibson PR . Histologic evidence of recurrent keratoconus seven years after keratoplasty. Am J Ophthalmol 1995; 119: 511–512.
Bechrakis N, Blom ML, Stark WJ, Green WR . Recurrent keratoconus. Cornea 1994; 13: 73–77.
Falls HF, Allen AW . Dominantly inherited keratoconus. J Genet Hum 1969; 17: 317–324.
Tyynismaa H, Sistonen P, Tuupanen S, Tervo T, Dammert A, Latvala T Alitalo . A locus for autosomal dominant keratoconus: linkage to 16q22.3-q23.1 in Finnish families. Invest Ophthalmol Vis Sci 2002; 43: 3160–3164.
Fullerton J, Paprocki P, Foote S, Mackey DA, Williamson R, Forrest S . Identity-by-descent approach to gene localization in eight individuals affected by keratoconus from north-west Tasmania. Australia Hum Genet 2002; 110: 462–470.
Hughes AE, Dash DP, Jackson AJ, Frazer DG, Silvestri G . Familiar keratoconus with cataract: linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest Ophthalmol Vis Sci 2003; 44: 5063–5066.
Brancati F, Valente EM, Sarkozy A, Feher J, Castori M, Del Duca P et al. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J Med Genet 2004; 41: 188–192.
Heon E, Greenberg A, Kopp KK, Rootman D, Vincent AL, Billingsley G et al. VSX1: a gene for posterior polymorphous dystrophy and keratoconus. Hum Mol Genet 2002; 11: 1029–1036.
Zhu LX, Yang HY, Wang YP, Figueroa KP, Rotter J, Pulst SM, Rabinowitz YS . Identification of a putative locus for keratoconus on Chromosome 21. Am J Hum Genet 1999; 65 ( Suppl.): 161.
Rabinowitz YS, Rasheed K, Yang H, Elashoff J . Accuracy of ultrasonic pachymetry and videokeratography in detecting keratoconus. J Cataract Refract Surg 1998; 24: 196–201.
Krachmer JH, Feder RS, Belin MW . Keratoconus and related non- inflammatory corneal thinning disorders. Surv Ophthalmol 1984; 28: 293–322.
Rabinowitz YS, Yang H, Akkina J, Brickman Y, Elashoff J, Rotter J . Videokeratography of normal human corneas. Br J Ophthalmol 1996; 80: 610–616.
Sczhotka L, Rabinowitz YS, Yang H . The influence of contact lens wear On the topography of keratoconus. CLAO J 1996; 22: 270–273.
Rasheed K, Rabinowitz YS, Remba M, Remba D . Inter and Intra-observer reliability of a classification scheme for corneal topographic patterns. Br J Ophthalmol 1998; 82: 1401–1406.
Sobel E, Lange K . Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 1996; 58: 1323–1337.
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES . Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996; 58: 1347–1363.
Terwilliger JD, Ott J . Handbook of Human Genetic Linkage. Baltimore, Md: Johns Hopkins University Press, 1994.
Acknowledgements
This research is supported by NIH NEI 09052, GCRC NIH 50091, and the Eye Birth Defects Research Foundation Inc. The initial genome scan was completed with the help of the General Clinical Research Center, Genotyping Core, grant no. MO1-RR00425.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tang, Y., Rabinowitz, Y., Taylor, K. et al. Genomewide linkage scan in a multigeneration Caucasian pedigree identifies a novel locus for keratoconus on chromosome 5q14.3-q21.1. Genet Med 7, 397–405 (2005). https://doi.org/10.1097/01.GIM.0000170772.41860.54
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1097/01.GIM.0000170772.41860.54
Keywords
This article is cited by
-
Association of KIF26B and COL4A4 gene polymorphisms with the risk of keratoconus in a sample of Iranian population
International Ophthalmology (2019)
-
The Proteins of Keratoconus: a Literature Review Exploring Their Contribution to the Pathophysiology of the Disease
Advances in Therapy (2019)