Abstract
As an indispensable molecular machine universal in all living organisms, the ribosome has been selected by evolution to be the natural target of many antibiotics and small-molecule inhibitors. High-resolution structures of pathogen ribosomes are crucial for understanding the general and unique aspects of translation control in disease-causing microbes. With cryo-electron microscopy technique, we have determined structures of the cytosolic ribosomes from two human parasites, Trichomonas vaginalis and Toxoplasma gondii, at resolution of 3.2–3.4 Å. Although the ribosomal proteins from both pathogens are typical members of eukaryotic families, with a co-evolution pattern between certain species-specific insertions/extensions and neighboring ribosomal RNA (rRNA) expansion segments, the sizes of their rRNAs are sharply different. Very interestingly, rRNAs of T. vaginalis are in size comparable to prokaryotic counterparts, with nearly all the eukaryote-specific rRNA expansion segments missing. These structures facilitate the dissection of evolution path for ribosomal proteins and RNAs, and may aid in design of novel translation inhibitors.
Similar content being viewed by others
Introduction
The ribosome is responsible for protein biosynthesis in all living organisms, and also plays diverse roles in regulating various cellular activities1,2,3,4. Nearly half of the naturally occurring antibiotics target prokaryotic 70S ribosomes to inhibit protein translation5. Eukaryotic ribosomes are also targets of many small-molecule inhibitors6. Given the essentiality of the ribosome and the abnormal activity of translation in many types of cancers, the ribosome-related processes have been emerging as targets for novel cancer drug development7. Over the past decades, an increasing number of high-resolution structures of the ribosomes from different species have been obtained (i.e., see8,9,10,11,12,13,14,15,16,17,18,19,20,21,22). Many of these structures are in complex with small-molecule inhibitors (i.e., see5,6,11,23,24,25,26). With these structures, diverse mechanisms of small molecule-based inhibition have been elucidated in atomic details. Therefore, high-resolution structures of specific ribosomes, especially those from human pathogens are urgently needed not only for the elaboration of their unique translation regulation components but also for the structure-based drug design to overcome the increasing health threat from drug-resistant pathogens.
Trichomonas vaginalis, a hydrogenosome-containing parasitic protist, causes trichomoniasis in human. It can infect both sexes and cause vaginitis in women and urethritis in men. Women infected with T. vaginalis during pregnancy could result in adverse pregnancy outcomes, and trichomoniasis patients have a high risk of HIV infection27. According to the related report from the World Health Organization, trichomoniasis has been listed as the most prevalent non-viral sexually transmitted disease worldwide. Besides its significance in human health, the cytosolic ribosome of this parasite itself shows many unique features28. Toxoplasma gondii, the causative agent of toxoplasmosis, is one of the most important Apicomplexan protozoa. It is estimated that one third of human population is chronically infected with this pathogen29. Infection by this parasite in pregnant women could result in severe outcomes (e.g., encephalitis and ocular disease in newborns)30. T. gondii may also cause catastrophic consequences to immuno-compromised population and to those infected with HIV31.
In the present study, we report near-atomic structures (3.2–3.4 Å) of the cytosol ribosomes isolated from these two human pathogens using cryo-electron microscopy (cryo-EM). While the structure of the 80S ribosome from T. gondii is, in general, similar to that of the model unicellular organism, Saccharomyces cerevisiae9, the T. vaginalis ribosome represents an extremely reduced version of typical eukaryotic ribosomes, with nearly all of the eukaryote-specific rRNA expansion segments (ES) missing. These data offer structural clues for understanding evolution of ribosomes across kingdoms, and more importantly, provide key information for structure-guided design of ribosome-targeting small-molecule inhibitors, as well as for functional characterization of eukaryotic rRNA ES and protein extensions.
Results
Overall structure of the T. vaginalis 80S ribosome
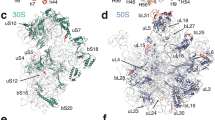
For the T. vaginalis 80S ribosome (Tv80S), we first obtained an overall map at a resolution of 3.30 Å (Supplementary information, Figures S1 and S2). To further improve the density maps for subunits, a soft mask-based refinement was applied, which resulted in two slightly improved maps for the small subunit (SSU) and large subunit (LSU), at the resolution of 3.35 Å and 3.20 Å, respectively (Supplementary information, Figure S2B). Most of the protein side chains and RNA bases are clearly separated in the SSU and LSU maps (Supplementary information, Figure S3). We were able to model 32 ribosomal proteins and about 95% of the rRNA residues (Figure 1A and 1B, Supplementary information, Table S1) with the T. vaginalis SSU map. Of the modeled proteins, all of them contain conserved globular domains, and about one third of them also have variable N-terminal or C-terminal extensions (NTE or CTE) compared with their yeast counterparts (Supplementary information, Table S1). We were only able to model a short C-terminal fragment (α-helix) of eS6 (Supplementary information, Table S1). Notably, phosphorylation of this C-terminus of eS6 is important in response to multiple physiological, pharmacological stimuli32. Protein eS31 located at the beak region was not modeled either, due to the flexibility of the beak in the SSU. Surprisingly, many rRNA ES (ES3S, ES6S, ES7S and ES12S) in typical eukaryotic SSUs that are expected to be in the foot and platform regions are largely absent in the Tv80S (Supplementary information, Figures S4-S7).
Overview and unique features of the T. vaginalis ribosome. (A, B) Atomic model of the Tv80S, shown in the front (A) and back (B) views. Ribosomal proteins are color-coded and rRNA molecules are shown in gray. (C) Cartoon comparison of the ribosomes from E. coli, T. vaginalis and S. cerevisieae, highlighting the hybrid composition of the T. vaginalis ribosome. (D) Evolutionary relationships of different species based on 16S-like rRNAs.
For the T. vaginalis LSU, we were able to model 41 ribosomal proteins and over 92% of the rRNA residues (Figure 1A and 1B, Supplementary information, Table S2). Around 40% of proteins have variable eukaryote-specific extensions. Five proteins, which locate at the L1 stalk (uL1) and P stalk (uL10, uL11, P1 and P2), were not modeled, as both the two stalks are known to be extremely flexible. Notably, we found that a rod-like density fits very well with the crystal structure of S. cerevisiae eL41 (Supplementary information, Figure S3B), but the primary sequence of eL41 is missing from the T. vaginalis protein database. Presumably, because the open reading frame of eL41 is less than 30 residues, it might have been filtered out of the gene prediction and annotation when the database was created33. Similar to the SSU, a unique feature of the T. vaginalis LSU is that nearly all of the rRNA ES (ES7L, ES9L, ES15L, ES19L, ES27L, ES31L, ES39L) are absent (Supplementary information, Figures S8–S10). Intriguingly, a highly flexible expansion segment ES27L in the bottom of the LSU, which mediates the interactions with various nascent polypeptide processing factors34, is also entirely gone in the Tv80S (Supplementary information, Figures S8 and S9E).
Altogether, the most intriguing finding is that the Tv80S typifies an unexpectedly “reduced” eukaryotic ribosome. Except for the presence of the 5.8S rRNA, the Tv80S appears to be a hybrid ribosome composed of a complete set of eukaryotic proteins and prokaryote-like 16S and 23S rRNAs (Figure 1C, Supplementary information, Figures S11 and S12). In addition to its outstanding divergence of the rRNAs in eukaryote-specific ES, certain rRNA helices of the Tv80S are even shorter than their E. coli counterparts8 (Supplementary information, Figure S9C-S9E). To determine whether the Tv80S resembles an ancient primitive eukaryote, we performed phylogenetic analysis of the SSU rRNAs from several representative species. Highly consistent with evolutionary analysis based on conserved ribosomal proteins28, our rRNA analysis suggests that T. vaginalis has branched very early (b1 in Figure 1D) from a certain evolutionary intermediate on the path to modern eukaryotes.
Overall structure of the T. gondii 80S ribosome
For the T. gondii 80S ribosome (Tg80S), a complete map was obtained at an overall resolution of 3.22 Å. “Focused” 3D refinement resulted in two improved maps for the SSU and LSU at the resolution of 3.35 Å and 3.18 Å, respectively (Supplementary information, Figures S13 and S14). In total, 32 and 42 ribosomal proteins were modeled for the SSU and LSU, respectively (Figure 2A and 2B, Supplementary information, Tables S3 and S4). Most of the rRNA residues were also successfully built. Similar to the Tv80S, five proteins of the LSU, which locate at the L1 stalk (uL1) and P stalk (uL10, uL11, P1, P2), were not modeled. Only one protein of the SSU, RACK1, which serves as a signaling scaffold35, was not modeled, because the local density of RACK1 was relatively weak. This observation is consistent with the fact that RACK1 is not always tightly associated with the ribosome36,37.
Overall structure of the T. gondii ribosome and species-specific features of the ES from LSU solvent surface view. (A, B) Atomic model of the Tg80S, shown in the front (A) and back (B) views. Ribosomal proteins are color-coded and rRNAs are in gray. (C-I) Eukaryotic rRNA ES, ES3S, ES7L and ES39L display species-specific features. For clarification, only three ES on the solvent surface are shown and the maps were low-pass filtered. The atomic models and density maps used are: (D) S. cerevisiae (PDB code: 4V88)9; (F) P. falciparum (PDB codes: 3J79 and 3J7A; EMDB code: 2661)11; (G) T. brucei (PDB code: 4V8M; EMDB code: 2239)13; (H) D. melanogaster (PDB code: 4V6W; EMDB code: 5591)14 and (I) H. sapiens (PDB code: 4V6X; EMDB code: 5592)14.
In general, structures of ribosomal proteins on the Tg80S are highly similar to those in other eukaryotic ribosomes. A notable example is eL28, which does not exist in the S. cerevisiae ribosome (Sc80S) but in other eukaryotic ribosomes, is also present in the Tg80S, although S. cerevisiae sits in the same sub-branch on the phylogenetic tree with T. gondii and its close relative P. falciparum (Figure 1D).
In contrast to ribosomal proteins, the rRNA ES of the Tg80S show many interesting dissimilarities with known structures of eukaryotic ribosomes. Overall, consistent with their evolutionary relationship (Figure 1D), structures of the ES on the Tg80S are more similar to those of the Sc80S and Pf80S11,36, especially for the ES on the SSU (Supplementary information, Figures S5–S7 and S15). However, the ES from the LSU appear to have evolved quite differently. For example, compared to the Sc80S and Pf80S, ES39L of the Tg80S is shortened, whereas its ES7L expands further in length to be the largest among the three (Figure 2D-2F, Supplementary information, Figures S16 and S17). Notably, these two ES of the human ribosome have significantly expanded and contain multiple helical branches (Figure 2I, Supplementary information, Figures S16F and S17G).
In short, the Tg80S structure is highly similar to other unicellular eukaryotic ribosomes, but with subtle differences in their rRNA ES.
Eukaryote-specific intersubunit bridges in the T. vaginalis 80S ribosome
Contacts between two ribosomal subunits are important for passing conformational signal during peptide elongation cycle and they play essential roles in regulating various aspects of translation9. Since most of the eukaryote-specific rRNA ES are absent in the Tv80S, we examined the regions of intersubunit bridges in the Tv80S. As a result, most of the central bridges that universally exist in both prokaryotic and eukaryotic ribosomes8,9,10 are well maintained in the Tv80S. Among the central bridges, eB14 is unique, because it is the only protein:RNA bridge in the center of the subunit interface. Bridge eB14 in the Tg80S is formed by the N-terminal protrusion of eL41 (a short, eukaryote-specific, single-helical protein) into the central pseudoknot region surrounded by h2, h27, h44, h45 and the joint of h19-h20 of the 18S rRNA (Supplementary information, Figure S18). This pseudoknot (h2) is a universal feature of the SSU in all domains of life38 and crucial for both the assembly and function of the SSU39. An interesting difference of eL41 in the Tg80S and Pf80S is the presence of an NTE (∼14 residues), which forms contract with h2 that are not seen in other ribosomes, such as the Sc80S9, Dm80S (D. melanogaster)14 and Hs80S (H. sapiens)14,15. Given the location of eL41 helix close to the decoding center and the presence of NTE in the central pseudoknot region (Supplementary information, Figure S18), this might reflect the subtle differences of ribosome biogenesis and decoding dynamics in the two parasites.
Apart from central bridges, many peripheral protein:RNA or protein:protein bridges display interesting differences. In the Sc80S and other eukaryotic ribosomes, ES31L (located at the base of L1 stalk) interacts with eS1 through two contact sites to form the bridge eB8 (Figure 3B), and substantial structural rearrangement of eB8 is seen among ribosomes in different conformational states9. With ES31L completely gone in the Tv80S (Supplementary information, Figure S9D), only one contact (L1:eS1) is retained (Figure 3A). For another example, the C-terminal α-helix of eL19 interacts with helix E in ES6S, and with protein eS7, to form bridge eB12 in the Sc80S and Hs80S (Figure 3E and 3F). However, this α-helix is much shorter (11 amino acids less) in the Tv80S (Figure 3D-3F, Supplementary information, Figure S19), and helix E of ES6S is also ∼30 nucleotides less than its S. cerevisiae counterpart (Supplementary information, Figure S4). Consequently, although all components involved in forming bridge eB12 in the Sc80S and Hs80S are present in the Tv80S, the shortened eL19 has lost its contact with eS7 (Figure 3D-3F).
Comparison of the eukaryote-specific bridges in the T. vaginalis, S. cerevisiae and H. sapiens ribosomes. (A-C) Zoomed-in view showing the bridge eB8 in the Tv80S (A), Sc80S (PDB code: 4v88; EMDB code: 6105)9,48 (B) and Hs80S (PDB code: 4V6X; EMDB code: 5592)14 (C). ES31L and L1 stalk are shown in red. (D-F) Same as A-C, but for the bridge eB12. The last α-helix of protein eL19 and rRNA ES6Sare markedly shorter in the Tv80S. Flexible components that were not modeled are indicated by dashed lines with the number of missing residues labeled. Length variation of eL19 is shown in the bottom cartoon.
In summary, through the analysis of the Tv80S in eukaryote-specific bridges, a general pattern is that many peripheral intersubunit bridges are still maintained, but with selected, specific contact sites lost, suggesting that the intersubunit association might be weakened in the Tv80S. This appears to be consistent with an early study showing that the T. vaginalis ribosomal factions were dominated by separate subunits under various buffer conditions, rather than monosomes in other species28.
Co-evolution of eL6, eL28, ES15L and ES7L
ES7L is one of the most diverse ES on the eukaryotic LSU. There is no ES7L in the Tv80S (Supplementary information, Figure S9C), as it has branched the evolutionary tree very early. The expansion of ES7L appears to occur in a step-wise fashion with increasing numbers of helical branches. As the largest ES in the Tg80S, ES7L consists three long helices, namely ES7La, ES7Lb and ES7Lc, stretching to the L1 stalk, the P stalk and CP regions, respectively (Figures 2E and 4D). Compared with the Sc80S, both branches of a and c of the Tg80S are significantly longer. In contrast, ES7Lfrom Trypanosomatida (such as T. brucei13, T. cruzi16 and L. donovani12,17) only has two branches (b and c; Figure 4C). This observation is consistent with the phylogenetic tree (Figure 1D), because Euglenozoa (Trypanosomatida is included in this group) has separated from the evolutionary stem earlier than the S. cerevisiae branch40. This indicates that the accretion of ES7La is an evolutionarily late event compared to the other two branches. In higher eukaryotes, such as H. sapiens12,14,15, ES7L has evolved to have as many as 7 branches (Figure 4G and Supplementary information, Figure S16F). Also consistently, the length expansion of ES15L(H45), which locates next to ES7L, correlates with the separation time of different species from the evolutionary tree, and reaches as long as 189 nucleotides in the Hs80S (Figure 4 and Supplementary information, Figure S20). A unique feature of the Tg80S is that lengthened tip of ES15L forms a set of base pairs (pseudoknot) with the insertion sequence (650-653) of ES7Lc (Supplementary information, Figure S21).
Co-evolution of eL6, eL28, ES15L and ES7L in different species. (A-G) Interactions of eL6 and eL28 with ES7L and ES15L in the ribosomes from different species. (A) T. vaginalis (lacking eL28), (B) S. cerevisiae (lacking eL28) (PDB code: 4v88)9, (C) T. brucei (PDB code: 4V8M)13, (D) T. gondii, (E) P. falciparum (PDB code: 3J79)11, (F) D. melanogaster (PDB code: 4V8W)14 and (G) H. sapiens (PDB code: 4V6X)14. Other ribosomal components were omitted for clarification. Branches of ES are labeled in lowercase letters, and protein termini are marked with N or C. Flexible components that were not modeled are indicated by dashed lines with the number of missing residues labeled.
Two proteins, eL28 and eL6, bind in the region of ES7L-ES15L, likely stabilizing the branches of these two ES (Figure 4 and Supplementary information, Figure S22). One interesting observation is that eL28 is not present in the Tv80S, and unlike other late separated species, the Sc80S does not have eL28 either. Given their evolutionary relationship, it is likely that eL28 is one of the lastly acquired eukaryote-specific ribosomal proteins, and the missing of eL28 in the Sc80S probably resulted from one ancient gene loss or inactivation. In addition, eL28 possesses a variable C-terminus, which could form diverse structures in different species, including both the loop and extended helix. This extension appears to have co-evolved with the complexity of the ES7La (Figure 4 and Supplementary information, Figure S22).
Protein eL6 is the best example to illustrate the co-evolution between ribosomal protein extensions and rRNA ES. The globular domain of eL6, which locates at the interface between ES7L and ES39L, is highly conserved in both sequence and structure (Figure 4 and Supplementary information, Figure S22). Notably, eL6 has an insertion in its globular domain (residues 122-127, T. gondii numbering), which is extremely diverse among different species. This insertion forms a lengthened protruding structure, contacting branch a of ES7L. By contrast, this insertion, as well as an additional stretch of sequences of eL6 (residues 101-143, S. cerevisiae numbering), is deleted in T. vaginalis (Supplementary information, Figure S22M), coincident with the total absence of ES7L in the Tv80S. Protein eL6 also contains a highly variable NTE (Supplementary information, Figure S22A-S22G), which differs markedly in both length and structure among different species. In the Tv80S, the NTE of eL6 extends with its terminal β-turn loop ending in a pocket formed by the helical base of H38, ES15L(H45), protein uL4 and uL30 (Supplementary information, Figure S23A). In contrast, in the Sc80S this NTE in fully extended form, makes extensive contacts with a different set of ribosomal components, including ES7L, ES15L, H46 and protein eL32 (Supplementary information, Figure S23B). In other eukaryotic ribosomes, the NTE of eL6 also takes very different path and makes contacts with distinct elements of ES7L and ES15L (Figure 4).
From the above examples, a general pattern is that the interactions between eukaryote-specific protein extensions/insertions and ribosomal components (including ES) are highly species-specific.
Discussion
Insights into the evolutionary path of ribosomal proteins and RNAs
In the present work, we report near-atomic structures of the cytosol ribosomes from two human pathogens, T. gondii and T. vaginalis. Very intriguingly, the Tv80S lacks almost all of the eukaryotic rRNA ES, despite the fact that T. vaginalis is still a member of eukaryotes28 and contain a full set of eukaryote-specific proteins. The sizes of the SSU and LSU rRNAs of the Tv80S is in fact comparable to those of bacterial rRNAs, except that the Tv80S also contains a 5.8S rRNA. Evolutionary analysis based on the SSU rRNA (Figure 1D) or ribosomal protein sequences indicates that T. vaginalis separated very early from the eukaryotic branch28,40, and therefore, retains many features of the primitive eukaryotic ribosome. This suggests that eukaryote-specific proteins have appeared much earlier than the rRNA ES. Eukaryotic ribosomal proteins were acquired progressively during evolution and eL28 is likely one of the latest proteins.
In terms of rRNA evolution, the ribosomes from Trypanosomatida (b2.1 in Figure 1D), including T. brucei13, T. cruzi16 and L. donovani12,17, are unique. Although they are, in general, similar to other unicellular eukaryotic ribosomes, their 28S rRNAs have further evolved to contain 6 segments, resulting from multiple specific cleavages during ribosome assembly. Consequently, as exemplified in the structures of leishmanial ribosomes, certain species-specific rRNA ES (such as ES42L) and trypanosomatida-specific protein extensions (such as uL13) play a role in stabilizing the ends of fragmented rRNAs12,17.
In summary, the two structures of the Tv80S and Tg80S, in combination with existing ribosome structures, provide a rich resource for understanding the co-evolution between rRNA ES and ribosomal protein extension/insertions.
T. gondii and T. vaginalis ribosomes as model systems for functional dissection of eukaryote-specific rRNA ES
The core of the ribosome is highly conserved and many aspects of canonical translation cycle have been understood at atomic details. However, little is known for the function of diverse rRNA ES seen in different eukaryotic species. With S. cerevisiae as a model system, it has been shown that many of the LSU ES (including ES39L, ES15L, ES9L and ES31L) are indispensable for cell growth and deletion of them causes various assembly defects41, indicating that many of the rRNA ES participate in ribosome biogenesis to varying extents. A hypothesis is that these ES play a scaffolding role in the assembly of eukaryote-specific proteins41. However, the Tv80S contains all eukaryotic ribosomal proteins but nearly no ES, thus challenging the idea that the primary roles of rRNA ES are in ribosome biogenesis.
On the other hand, ES in eukaryote ribosomes are generally believed to correlate with the complexity in translation regulation. However, far less is known for their potential roles in translation. The natural deficiency of the Tv80S in ES, therefore, allows an informative comparison with other eukaryotic species. In the Tv80S, both ES6S and ES7S, which interact with eIF3 complex in S. cerevisiae and mammalian systems42,43,44, are shortened (Supplementary information, Figures S6 and S7). Thus, their contacts with initiation machinery should be gone. Notably, the eIF3 complex in S. cerevisiae contain six subunits (eIF3a, eIF3b, eIF3c, eIF3g, eIF3i and eIF3j), whereas in mammals, the number of subunits increases to 13 (a-m). Bioinformatics survey of the T. vaginalis proteome only identified eIF3b, eIF3c, eIF3d, eIF3h and eIF3i, but not the core subunit eIF3a45. The failure might result from the low sequence homology of the translation components between T. vaginalis and other eukaryotes. Nevertheless, the extremely shortened ES6S and ES7S in the Tv80S suggests that the interplay between the eIF3 complex and SSU in T. vaginalis might be unique to a certain extent.
For another example, in the Tv80S, ES27L is totally missing and ES12S is also shortened (Supplementary information, Figure S9E). ES27L is the largest expansion segment in the Sc80S. It is indispensable for viability in S. cerevisiae41 and in Tetrahymena thermophila46. Both ES27L and ES12S interact with Zuotin (a J-protein from the HSP40 family), a component of the co-translational chaperone system in S. cerevisiae47,48,49. By binding to the two ES from the LSU and SSU, Zuotin virtually creates a rigid bridge between the two subunits48. Zuotin partners with Ssz to form the ribosome-associated complex (RAC), which together with Ssb facilitates the early folding of nascent peptides emerging from the peptide exit tunnel. Consistent with the structural differences in ES27L and ES12S, we did not find Zuotin homologue in the T. vaginalis genome. Of all proteins that belong to J-protein family in T. vaginalis, we could not locate anyone that contains the reported highly conserved residues required for the function of RAC49. Presumably, the co-translational RAC does not exist in the T. vaginalis system. Therefore, it appears that the sizes of ES27L and ES12S correlate with the complexity of the co-translational chaperone system.
ES7L is one of the most variable regions in the eukaryotic rRNA. Deletion of this segment results in growth defect41. A previous study showed that the fragment of ES7L from S. cerevisiae could bind in vitro to proteins involved in different cellular processes, such as cellular response to abiotic stimulus, LSU biogenesis and amino acid synthesis50. A sharp contrast is that ES7Lis completely truncated in T. vaginalis, but significantly enlarged in T. gondii and grows into a gigantic complex structure in metazoan (Figure 2E-2I and Supplementary information, Figure S16). These differences may reflect species-specific coupling of translation with other cellular regulatory pathways. In fact, T. gondii has a very complicate life cycle between the definite host (feline animals such as cats) and intermediate hosts (all warm-blooded animals including humans), whereas T. vaginalis presents a very simple life cycle with only a trophozoite by binary division in human vagina.
Nevertheless, the specific functions of individual rRNA ES in protein translation and/or ribosome biogenesis remain to be experimentally explored. Our findings on the Tv80S suggest that a comparative study using both T. vaginalis and S. cerevisiae may be particularly useful.
Species-specific roles of eukaryotic ribosomal protein extensions/insertions
Similar to the rRNAs, a feature of eukaryotic ribosomal proteins is that many of them have N- and/or C-terminal extensions, as well as insertions on their conserved globular domains. These sequences vary strikingly across species. Using the S. cerevisiae system, functions of some of the eukaryote-specific extensions/insertions have been explored (e.g., see51,52,53,54,55,56,57,58,59,60). These studies have revealed a collection of functions: harboring nuclear localization signal, rRNA processing, subunit assembly and recruiting certain translational components (54 and reference therein).
Based on the above-mentioned data, our structural models enable a cross-species examination of eukaryote-specific extensions/insertions of ribosomal proteins. An interesting observation is that many extensions may have species-specific roles. Taking uL4 as an example, it has a very long C-terminal extension (Supplementary information, Figure S24). In both Tv80S and Tg80S, this extension (in different secondary structures) wanders on the solvent surface of the LSU, with the terminal portion interacting with uL16 (Figure 5A and 5B), whereas the human uL4, with the longest CTE though, has no such a contact with uL16 (Figure 5D). The CTE of the yeast uL4 is the shortest, and is not able to reach the position of uL16 (Figure 5C). It was shown that depletion of the full-length uL4 in S. cerevisiae blocks the LSU biogenesis at early steps, however, truncation of only the CTE of uL4 does not cause apparent assembly defects53,61. Protein uL16 is a late binding protein, and its incorporation into the peptidyl transferase center constitutes a quality control checkpoint of the LSU assembly62,63. Therefore, the species-specific interaction between uL4-CTE with uL16 suggests that certain late steps of ribosome biogenesis in the two parasites might be different. For another instance, uS3 has a variable CTE, which display a species-specific interaction with RACK1. By contrast, in the Tv80S, this CTE is much shorter and not able to touch RACK1 (Supplementary information, Figure S25), suggesting a possible difference of the Tv80S in RACK1-mediated cellular signaling35.
CTE of uL4 makes species-specific interaction with uL16. (A) The long CTE of uL4 interacts with uL16 in the Tv80S. The N- and C-termini of uL4 are labeled as N and C, respectively. (B-D) Same as A, but for the corresponding components of the Tg80S, Sc80S and Hs80S. Atomic models and maps used are: (C) S. cerevisiae (PDB code: 4V88)9, (D) H. sapiens (PDB code: 4V6X; EMDB code: 5592)14. The middle inset shows the comparison of uL4-CTE from T. vaginalis and H. sapiens.
Implication in anti-parasite drug design
As a member of amitochondrial protozoan, T. vaginalis makes use of hydrogenosome to produce ATP. The widely-used drug metronidazole targets this specific organelle64. However, resistance to the nitroimidazole class of drugs has been increasingly found. In the treatment of toxoplasmosis, dihydrofolate reductase and dihydropteroate synthase are two main targets, but the host toxicity is also rather high. A few classes of ribosome-targeting drugs were also tested on T. vaginalis and T. gondii. However, many of them also have severe side effects to the host. Among these drugs, the selectivity of paramomycin (a natural aminoglycoside) on leishmanial ribosomes over mammalian ribosomes65 have been explored by structural approaches12,66. These studies suggested that the presence of non-canonical base pairing in the decoding center (E. coli numbering, 1 406-1 409 and 1 491-1 495) and the resulting subtle structural difference of various ribosomes dictate the specificity of the drug12,66. Specifically, the Watson-Crick base pair of C1409:G1491 of bacterial ribosomes10 or the equivalent U:A base pair of trypanosomatidal ribosomes is replaced by a non-canonical C:A pair in the human cytosol ribosome. This base pair is important in stacking with one ring of paromomycin and the alteration in the Hs80S may decrease the affinity of paromomycin12. In both Tv80S and Tg80S, the equivalent positions of these two residues are identical to bacterial ones, capable of forming canonical base pair (Supplementary information, Figure S26). This suggest that paromomycin could efficiently target both T. vaginalis and T. gondii ribosomes. Consistent with this prediction, many cases of trichomoniasis have been reported to be cured successfully with paromomycin67.
Nevertheless, because the functional core of the ribosome is highly conserved, traditional inhibitors targeting the decoding center, peptidyl transferase center, and the polypeptide exit tunnel are usually rather broad-spectrum5, and would inevitably introduce undesired side effects to the host cell. Since the eukaryote-specific rRNA ES and protein extensions are mostly species-specific, and parasite ribosomes may differ very largely in ribosome biogenesis, inhibitors that target ribosome biogenesis68 or unique translation regulatory circuits of parasites69 might ideally be able to provide excellent specificity to particular disease-causing microbes.
Materials and Methods
Isolation of cytosolic ribosomes from T. vaginalis and T. gondii
T. vaginalis trophozoites (GZTv21 strain) were cultured in TYM medium70, and were harvested by centrifugation at 1 000× g for 10 min during log phase. Tachyzoites of T. gondii (RH strain) were maintained by serial passage in human foreskin fibroblast (HFF) cell monolayer grown in Dulbecco's modified Eagle medium (DMEM, HyClone) supplemented with 10% (V/V) fetal bovine serum (Gibco). After full lysis of infected HFFs, tachyzoites were harvested and passed through 27-g needle for three times. Then, tachyzoite pellets were collected by centrifugation at 1 000× g for 10 min. For both T. vaginalis and T. gondii, cell pellets were washed with PBS and freeze-thawed five times in ribosome buffer (25 mM HEPES, pH 7.4, 400 mM KAcO, 15 mM Mg(AcO)2, 1 mM DTT) containing 0.5% Triton X-100 and protease inhibitor cocktail. The lysate was centrifuged at 20 000× g for 30 min and the supernatant was loaded on the top of 1.25 mL cushion buffer (25 mM HEPES-KOH, pH 7.4, 400 mM KAcO, 15 mM Mg(AcO)2, 1 mM DTT, 0.5 M sucrose) and centrifuged at 48 000 rpm for 4 h in a MLS50Ti rotator (Beckman Coulter). The pellet was re-suspended in ribosome buffer and 80S ribosomes were separated by centrifugation through a 10%-40% sucrose gradient in ribosome buffer.
Molecular phylogenetic analysis based on 16S-like rRNA
For the analysis of evolution relationships, software MEGA7 was used71. SSU rRNAs of different species were aligned using the built-in Clustal. Then the Neighbor-Joining method72,73 was used to infer the phylogenetic tree based on the aligned 16S/18S rRNA sequences. Another built-in maximum likelihood method based on the Tamura-Nei model74 was used to validate the phylogenetic tree. Finally, the optimal tree with the sum of branch length = 1.96377475 is shown (Figure 1D). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method73 and are in the units of the number of base substitutions per site. The analysis involved 10 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1 167 positions in the final dataset.
Cryo-EM data acquisition
3.5-μL aliquots of purified ribosomes at a concentration of ∼100 nM were added to glow-discharged holey carbon grids (Quantifoil R2/2) on which a continuous thin carbon film (estimated ∼40 Å) was deposited. Grids were blotted for 2.5 s with an FEI Vitrobot Mark IV (100% humidity, 4 °C), and then plunged into liquid ethane immediately. Grids were transferred to an FEI Titan Krios electron microscope for data collection (operated at 300 kV). Image stacks were recorded on a Gatan K2 Summit direct electron detector in its counting mode with a nominal magnification of 22 500×, corresponding to a pixel size of 1.32 Å at the object scale. The defocus varied from −1.6 to −2.5 μm. All image stacks were collected using UCSF Image475 under low-dose conditions. Each image stack contains 32 dose-fractionated frames with a dose rate of ∼8 counts per physical pixel per second for a total exposure time of 8 s.
Image processing
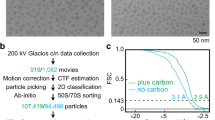
For T. vaginalis ribosomal particles, the dose-fractionated frames of each raw stack were aligned and summed to generate a final micrograph using MOTIONCORR76. EMAN277 was used for manual particle-picking and micrograph-screening. The contrast transfer function (CTF) parameters of each micrograph were estimated using CTFFIND378. The 2D and 3D classification and refinement were carried out with RELION79. Reference-free 2D classification was used to screen particles (Supplementary information, Figure S1A). Particles with good 2D class averages were selected for the following 3D classification into 8 classes (Supplementary information, Figure S1B). A density map of S. cerevisiae 80S ribosome, which was generated from the crystal structure (PDB code: 4v88), was used as initial model (low-pass filtered to 40 Å). Except for two classes (∼38% of all particles) of the LSU, structures of other groups revealed ribosomes in different conformations (Supplementary information, Figure S1B). One class (∼8.4%) represents a classical non-rotated state, whereas the other four belong to differently ratcheted states. Among the five good classes, one or two tRNAs were found in class structures of the Tv80S. Class 3 and Class 7 contain two tRNAs, one at the A/P site and the other at the P/E site, but the relative rotations between head and body are slightly different in these two classes. Class 8 has classical A-site and P-site tRNAs, but the P-site tRNA is relatively weak. Class 5 and Class 6 only have one P-site tRNA, and they are highly similar. We combined Class 5 and Class 6 (25.5%) for further 3D refinement (Supplementary information, Figure S1B). To reduce the possible radiation damage, only frames 2 to 15 of each raw stack were selected for creating the dose-reduced micrograph. The new particles extracted from the dose-reduced micrographs were subjected to 3D refinement. Furthermore, to estimate the CTF parameters more accurately, local refinement of CTF parameters was performed using Gctf80. The CTF parameters in the input particle star file that was used for 3D refinement were replaced by the newly estimated ones. After the convergence of 3D refinement using dose-reduced particles and refined CTF parameters, additional rounds of “focused” 3D refinement were applied by adding soft-edged masks of the whole 80S ribosome, the LSU or SSU. The soft masks for the SSU and LSU were made as follows. First, the density maps of the SSU and LSU were segmented out of the whole ribosome map using UCSF Chimera81. The segmented maps were then converted into binary maps. The [0-1] sharp edges in the binary maps were “softened” by adding cosine-shaped edges with RELION to create the soft masks. As a result, the density map of the whole ribosome converged at an overall resolution of 3.3 Å, with local resolution of the LSU and SSU regions at 3.24 Å and 3.7 Å, respectively. Application of the soft masks of individual subunits improved the maps of the LSU and SSU to a resolution of 3.2 Å and 3.35 Å, respectively.
The T. gondii ribosomal particles were processed following the same procedures (Supplementary information, Figure S13). Of the eight classes from 3D classification, seven of them (∼92.8%) represent differently rotated structures (with a very weak E-site tRNA). Subsequently, a complete density map for the Tg80S, derived from particles of four similarly rotated classes (∼63.3%), was obtained at an overall resolution of 3.22 Å. Focused 3D refinement improved the resolution from 3.2 Å to 3.18 Å for the LSU and from 3.5 Å to 3.35 Å for the SSU (Supplementary information, Figure S14). All the resolution estimations indicated above were based on the gold-standard FSC 0.143 criterion with a soft mask applied to half maps, and the mask effect was corrected by RELION82. Local resolution variations were estimated using ResMap83.
Model building and refinement
Crystal structure of the S. cerevisiae 80S ribosome (PDB codes: 3U5B, 3U5C, 3U5D and 3U5E) was used as the initial template for modeling of both the T. vaginalis and T. gondii ribosomes. At first, individual chains of rRNAs and r-proteins from the crystal structure of the S. cerevisiae ribosome were docked into the density maps of the T. vaginalis and T. gondii ribosomes as rigid bodies using UCSF Chimera.
For the modeling of the r-proteins, their sequences were obtained from the Uniprot database (http://www.uniprot.org/) and aligned with their corresponding proteins in S. cerevisiae using Clustal Omega84 to analyze the conserved regions and extension differences. The globular domains of r-proteins are conserved for both T. vaginalis and T. gondii, except that eL28 is only present in the T. gondii ribosome, but missing from both the S. cerevisiae and T. vaginalis ribosomes. For each r-protein, extensive residue replacement and model rebuilding were done manually using COOT85.
For building of rRNAs, sequences were obtained from SILVA86 and aligned with the yeast rRNAs using Clustal Omega84. Conserved regions/fragments between the T. vaginalis and S. cerevisiae rRNAs, and between the T. gondii and S. cerevisiae rRNAs were chosen as starting points for the modeling. Specifically, these fragments of the S. cerevisiae 80S crystal structure were docked into the density map and individual rRNA residues were mutated based on the sequences of the T. vaginalis or T. gondii versions. Less conserved regions and linker sequences were built de novo using COOT. To build the model of the T. gondii ribosome, rRNA secondary structural map downloaded from the Comparative RNA Website was used to aid the RNA modeling. Base-pairs were reviewed after manual adjustment with COOT85.
The atomic model refinements and cross-validations of the SSU and LSU of T. vaginalis and T. gondii ribosomes were done subsequently. The atomic model was first refined against the corresponding density map by real-space refinement (phenix.real_space_refine)87 in PHENIX88. After refinement, alternating rounds of manual model adjustment using COOT and model refinement using PHENIX were applied. Further refinement was carried out using REFMAC89 in Fourier space according to previously established protocols90 with secondary structure and geometry constraints applied. Different weights of density maps for refinement were tested. Cross-validation against overfitting was performed following the procedures previously described90. The final model was evaluated using MolProbity (Supplementary information, Tables S5 and S6).
Accession codes
The cryo-EM density maps of the Tv60S, Tv40S, Tg60S and Tg40S have been deposited in the EMDB with accession numbers of EMD-6784, EMD-6788, EMD-6778 and EMD-6780, respectively. The atomic models have been deposited in the PDB with accession numbers of 5XY3, 5XYI, 5XXB and 5XXU, respectively.
Author Contributions
NG designed and directed experiments. ZFL and QG purified the ribosome samples, collected cryo-EM data, performed image processing and analyzed structures. ZFL and LZ performed structural modeling. XS and YJ cultured the T. gondii tachyzoites. YTX, DHL and ZRL cultured T. vaginalis trophozoites. ZFL and NG wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
Inada T . The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem Sci 2017; 42:5–15.
Farley KI, Baserga SJ . Probing the mechanisms underlying human diseases in making ribosomes. Biochem Soc Trans 2016; 44:1035–1044.
Buszczak M, Signer RA, Morrison SJ . Cellular differences in protein synthesis regulate tissue homeostasis. Cell 2014; 159:242–251.
Harper JW, Bennett EJ . Proteome complexity and the forces that drive proteome imbalance. Nature 2016; 537:328–338.
Wilson DN . Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 2014; 12:35–48.
Garreau de Loubresse N, Prokhorova I, Holtkamp W, et al. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014; 513:517–522.
Ruggero D . Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013; 5.
Schuwirth BS, Borovinskaya MA, Hau CW, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science 2005; 310:827–834.
Ben-Shem A, Garreau de Loubresse N, Melnikov S, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 2011; 334:1524–1529.
Selmer M, Dunham CM, Murphy FV 4th, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006; 313:1935–1942.
Wong W, Bai XC, Brown A, et al. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 2014; 3.
Zhang X, Lai M, Chang W, et al. Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes. Nat Commun 2016; 7:13223.
Hashem Y, des Georges A, Fu J, et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 2013; 494:385–389.
Anger AM, Armache JP, Berninghausen O, et al. Structures of the human and Drosophila 80S ribosome. Nature 2013; 497:80–85.
Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP . Structure of the human 80S ribosome. Nature 2015; 520:640–645.
Liu Z, Gutierrez-Vargas C, Wei J, et al. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc Natl Acad Sci USA 2016; 113:12174–12179.
Shalev-Benami M, Zhang Y, Matzov D, et al. 2.8-Å cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Rep 2016; 16:288–294.
Desai N, Brown A, Amunts A, Ramakrishnan V . The structure of the yeast mitochondrial ribosome. Science 2017; 355:528–531.
Greber BJ, Bieri P, Leibundgut M, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 2015; 348:303–308.
Amunts A, Brown A, Toots J, Scheres SH, Ramakrishnan V . Ribosome. The structure of the human mitochondrial ribosome. Science 2015; 348:95–98.
Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N . The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 2017; 36:475–486.
Ahmed T, Yin Z, Bhushan S . Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep 2016; 6:35793.
Brodersen DE, Clemons WM, Carter AP, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000; 103:1143–1154.
Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA . The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol 2010; 17:289–293.
Schlunzen F, Zarivach R, Harms J, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001; 413:814–821.
Wilson DN, Schluenzen F, Harms JM, et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA 2008; 105:13339–13344.
Leitsch D . Recent advances in the Trichomonas vaginalis Field. F1000Res 2016; 5.
Arisue N, Maki Y, Yoshida H, et al. Comparative analysis of the ribosomal components of the hydrogenosome-containing protist, Trichomonas vaginalis. J Mol Evol 2004; 59:59–71.
Pappas G, Roussos N, Falagas ME . Toxoplasmosis snapshots: global status of Toxoplasma gondii seroprevalence and implications for pregnancy and congenital toxoplasmosis. Int J Parasitol 2009; 39:1385–1394.
Fatoohi AF, Cozon GJ, Greenland T, et al. Cellular immune responses to recombinant antigens in pregnant women chronically infected with Toxoplasma gondii. Clin Diagn Lab Immunol 2002; 9:704–707.
Lau YL, Lee WC, Gudimella R, et al. Deciphering the draft genome of Toxoplasma gondii RH strain. PLoS One 2016; 11:e0157901.
Ruvinsky I, Meyuhas O . Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 2006; 31:342–348.
Carlton JM, Hirt RP, Silva JC, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 2007; 315:207–212.
Melnikov S, Ben-Shem A, Garreau de Loubresse N, et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 2012; 19:560–567.
Nilsson J, Sengupta J, Frank J, Nissen P . Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 2004; 5:1137–1141.
Sun M, Li W, Blomqvist K, et al. Dynamical features of the Plasmodium falciparum ribosome during translation. Nucleic Acids Res 2015; 43:10515–10524.
Coyle SM, Gilbert WV, Doudna JA . Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 2009; 29:1626–1634.
Petrov AS, Bernier CR, Gulen B, et al. Secondary structures of rRNAs from all three domains of life. PloS One 2014; 9:e88222.
Sardana R, Liu X, Granneman S, et al. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PloS Biol 2015; 13:e1002083.
Sogin ML, Silberman JD . Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int J Parasitol 1998; 28:11–20.
Ramesh M, Woolford JL, Jr. Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 2016; 22:1153–1162.
des Georges A, Dhote V, Kuhn L, et al. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 2015; 525:491–495.
Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N . Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol 2015; 22:269–271.
Erzberger JP, Stengel F, Pellarin R, et al. Molecular architecture of the 40SeIF1eIF3 translation initiation complex. Cell 2014; 158:1123–1135.
Rezende AM, Assis LA, Nunes EC, et al. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates — identification of conserved and divergent features based on orthologue analysis. BMC Genomics 2014; 15:1175.
Sweeney R, Chen L, Yao MC . An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol 1994; 14:4203–4215.
Leidig C, Bange G, Kopp J, et al. Structural characterization of a eukaryotic chaperone — the ribosome-associated complex. Nat Struct Mol Biol 2013; 20:23–28.
Zhang Y, Ma C, Yuan Y, et al. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat Struct Mol Biol 2014; 21:1042–1046.
Lee K, Sharma R, Shrestha OK, Bingman CA, Craig EA . Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits. Nat Struct Mol Biol 2016; 23:1003–1010.
Gomez Ramos LM, Smeekens JM, Kovacs NA, et al. Yeast rRNA expansion segments: folding and function. J Mol Biol 2016; 428:4048–4059.
Jakovljevic J, Ohmayer U, Gamalinda M, et al. Ribosomal proteins L7 and L8 function in concert with six A(3) assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 2012; 18:1805–1822.
Tutuncuoglu B, Jakovljevic J, Wu S, Gao N, Woolford JL Jr. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. RNA 2016; 22:1386–1399.
Gamalinda M, Woolford JL Jr. Deletion of L4 domains reveals insights into the importance of ribosomal protein extensions in eukaryotic ribosome assembly. RNA 2014; 20:1725–1731.
Fernandez-Pevida A, Martin-Villanueva S, Murat G, et al. The eukaryote-specific N-terminal extension of ribosomal protein S31 contributes to the assembly and function of 40S ribosomal subunits. Nucleic Acids Res 2016; 44:7777–7791.
Pillet B, Garcia-Gomez JJ, Pausch P, et al. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet 2015; 11:e1005565.
Stelter P, Huber FM, Kunze R, et al. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol Cell 2015; 58:854–862.
Lumsden T, Bentley AA, Beutler W, et al. Yeast strains with N-terminally truncated ribosomal protein S5: implications for the evolution, structure and function of the Rps5/Rps7 proteins. Nucleic Acids Res 2010; 38:1261–1272.
Garcia-Gomez JJ, Fernandez-Pevida A, Lebaron S, et al. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet 2014; 10:e1004205.
Kressler D, Bange G, Ogawa Y, et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 2012; 338:666–671.
Pratte D, Singh U, Murat G, Kressler D . Mak5 and Ebp2 act together on early pre-60S particles and their reduced functionality bypasses the requirement for the essential pre-60S factor Nsa1. PloS One 2013; 8:e82741.
Gamalinda M, Ohmayer U, Jakovljevic J, et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 2014; 28:198–210.
Sulima SO, Gulay SP, Anjos M, et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res 2014; 42:2049–2063.
Ma C, Wu S, Li N, et al. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 2017; 24:214–220.
Vieira PD, Giordani RB, Macedo AJ, Tasca T . Natural and synthetic compound anti-Trichomonas vaginalis: an update review. Parasitol Res 2015; 114:1249–1261.
Fernandez MM, Malchiodi EL, Algranati ID . Differential effects of paromomycin on ribosomes of Leishmania mexicana and mammalian cells. Antimicrob Agents Chemother 2011; 55:86–93.
Shalev M, Rozenberg H, Smolkin B, et al. Structural basis for selective targeting of leishmanial ribosomes: aminoglycoside derivatives as promising therapeutics. Nucleic Acids Res 2015; 43:8601–8613.
Stover KR, Riche DM, Gandy CL, Henderson H . What would we do without metronidazole? Am J Med Sci 2012; 343:316–319.
Stokes JM, Brown ED . Chemical modulators of ribosome biogenesis as biological probes. Nat Chem Biol 2015; 11:924–932.
Joyce BR, Konrad C, Wek RC, Sullivan WJ . Translation control is critical during acute and chronic stages of toxoplasmosis infection. Expert Rev Anti-Infe 2011; 9:1–3.
Diamond LS . The establishment of various trichomonads of animals and man in axenic cultures. J Parasitol 1957; 43:488–490.
Kumar S, Stecher G, Tamura K . MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870–1874.
Saitou N, Nei M . The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–425.
Tamura K, Nei M, Kumar S . Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 2004; 101:11030–11035.
Tamura K, Nei M . Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10:512–526.
Li X, Zheng S, Agard DA, Cheng Y . Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol 2015; 192:174–178.
Li X, Mooney P, Zheng S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013; 10:584–590.
Tang G, Peng L, Baldwin PR, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 2007; 157:38–46.
Mindell JA, Grigorieff N . Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347.
Scheres SH . RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530.
Zhang K . Gctf: Real-time CTF determination and correction. J Struct Biol 2016; 193:1–12.
Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612.
Chen S, McMullan G, Faruqi AR, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 2013; 135:24–35.
Kucukelbir A, Sigworth FJ, Tagare HD . Quantifying the local resolution of cryo-EM density maps. Nat Methods 2014; 11:63–65.
Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7:539.
Emsley P, Lohkamp B, Scott WG, Cowtan K . Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66:486–501.
Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41:D590–D596.
Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D 2012; 68:352–367.
Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 2010; 66:213–221.
Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 1997; 53:240–255.
Amunts A, Brown A, Bai XC, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014; 343:1485–1489.
Acknowledgements
We thank the Tsinghua University Cryo-EM Facility of China National Center for Protein Sciences (Beijing) for providing resources for data collection and computation. Part of the computation was performed on the Computing Platform of the Center for Life Science, Peking University. This work was supported by the National Natural Science Foundation of China (31630087, 31422016 and 31470722 to NG; 31472058 and 31672276 to ZRL) and by the Special Fund for Agro-scientific Research in the Public Interest (201303042 to XS).
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Cryo-EM data processing of the T. vaginalis ribosome particles. (PDF 677 kb)
Supplementary information, Figure S2
Resolution estimation and model validation of the final T. vaginalis ribosomal maps. (PDF 628 kb)
Supplementary information, Figure S3
Representative densities of different elements in the density map of the T. vaginalis ribosome. (PDF 447 kb)
Supplementary information, Figure S4
Comparison of secondary structure diagram of the SSU rRNA between T. vaginalis and S. cerevisiae. (PDF 391 kb)
Supplementary information, Figure S5
Comparison of ES3S among selected representative species. (PDF 274 kb)
Supplementary information, Figure S6
Comparison of ES6S among selected representative species. (PDF 274 kb)
Supplementary information, Figure S7
Comparison of ES7S among selected representative species. (PDF 250 kb)
Supplementary information, Figure S8
Comparison of secondary structure diagram for the LSU rRNA between T. vaginalis and S. cerevisiae. (PDF 502 kb)
Supplementary information, Figure S9
Comparison of selected representative rRNA helices between T. vaginalis ribosome and E. coli ribosome. (PDF 230 kb)
Supplementary information, Figure S10
Comparison of ES31L among selected representative species. (PDF 231 kb)
Supplementary information, Figure S11
T. vaginalis ribosome is a “hybrid” of prokaryote-like rRNA core and eukaryotic r-proteins shell. (PDF 507 kb)
Supplementary information, Figure S12
Comparison of the rRNA between T. vaginalis ribosome and E. coli ribosome. (PDF 473 kb)
Supplementary information, Figure S13
Cryo-EM data processing of the T. gondii ribosome particles. (PDF 635 kb)
Supplementary information, Figure S14
Resolution estimation and model validation of the final T. gondii ribosomal maps. (PDF 679 kb)
Supplementary information, Figure S15
Comparison of selected rRNA expansion segments on the SSU from a few representative species. (PDF 775 kb)
Supplementary information, Figure S16
Comparison of ES7L among selected representative species. (PDF 265 kb)
Supplementary information, Figure S17
Comparison of ES39L among selected representative species. (PDF 192 kb)
Supplementary information, Figure S18
Comparison of Bridge eB14 in selected representative species. (PDF 276 kb)
Supplementary information, Figure S19
Sequence alignment of eL19 from T. vaginalis, S. cerevisiae and H. sapiens. (PDF 252 kb)
Supplementary information, Figure S20
Comparison of ES15L among selected representative species. (PDF 187 kb)
Supplementary information, Figure S21
Secondary structure diagram for the prolonged ES7L in the T. gondii LSU rRNA and pseudo-knot between ES7L and ES15L. (PDF 241 kb)
Supplementary information, Figure S22
Comparison of protein eL6 and eL28 from selected representative species. (PDF 1246 kb)
Supplementary information, Figure S23
Species-specific extension of eL6 interacts with different components. (PDF 259 kb)
Supplementary information, Figure S24
Species-specific ribosomal protein extensions of uL4. (PDF 823 kb)
Supplementary information, Figure S25
Species-specific interaction between uS3 and RACK1. (PDF 533 kb)
Supplementary information, Figure S26
Sequence comparison of the decoding centers (h44) on the SSU from a few pathogenic species. (PDF 703 kb)
Supplementary information, Table S1
Ribosomal proteins and rRNAs of the T. vaginalis SSU (XLSX 13 kb)
Supplementary information, Table S2
Ribosomal proteins and rRNAs of the T. vaginalis LSU (XLSX 15 kb)
Supplementary information, Table S3
Ribosomal proteins and rRNAs of the T. gondii SSU (XLSX 13 kb)
Supplementary information, Table S4
Ribosomal proteins and rRNAs of the T. gondii LSU (XLSX 17 kb)
Supplementary information, Table S5
Statistics of data collection and model refinement of the T. vaginalis ribosome (PDF 127 kb)
Supplementary information, Table S6
Statistics of data collection and model refinement of the T. gondii ribosome. (PDF 126 kb)
Rights and permissions
About this article
Cite this article
Li, Z., Guo, Q., Zheng, L. et al. Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res 27, 1275–1288 (2017). https://doi.org/10.1038/cr.2017.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2017.104
Keywords
This article is cited by
-
Structures of the eukaryotic ribosome and its translational states in situ
Nature Communications (2022)
-
Evolution of ribosomal protein network architectures
Scientific Reports (2021)