Abstract
Interferons (IFNs) are crucial for host defence against viruses. Many IFN-stimulated genes (ISGs) induced by viral infection exert antiviral effects. Microarray analysis of gene expression induced in liver tissues of mice on dengue virus (DENV) infection has led to identification of the ISG gene ISG12b2. ISG12b2 is also dramatically induced on DENV infection of Hepa 1-6 cells (mouse hepatoma cell line). Here, we performed biochemical and functional analyses of ISG12b2. We demonstrate that ISG12b2 is an inner mitochondrial membrane (IMM) protein containing a cleavable mitochondrial targeting sequence and multiple transmembrane segments. Overexpression of ISG12b2 in Hepa 1-6 induced release of cytochrome c from mitochondria, disruption of the mitochondrial membrane potential, and activation of caspase-9, caspase-3, and caspase-8. Treatment of ISG12b2-overexpressing Hepa 1-6 with inhibitors of pan-caspase, caspase-9, or caspase-3, but not caspase-8, reduced apoptotic cell death, suggesting that ISG12b2 activates the intrinsic apoptotic pathway. Of particular interest, we further demonstrated that ISG12b2 formed oligomers, and that ISG12b2 was able to mediate apoptosis through both Bax/Bak-dependent and Bax/Bak-independent pathways. Our study demonstrates that the ISG12b2 is a novel IMM protein induced by IFNs and regulates mitochondria-mediated apoptosis during viral infection.
Similar content being viewed by others
Main
Type I interferons (IFNs) are the best-known molecules critical for the resistance to viral infection and the regulation of immune response in host defence against viral infection.1 Type I IFNs mediate antiviral effects through the action of various IFN-stimulated genes (ISGs); investigating the functions of these gene products is therefore important for understanding the molecular mechanisms underlying the antiviral action of type I IFNs. Although >300 ISGs have been identified, only a few ISGs involved in antiviral effects have been defined,2, 3 including dsRNA-activated protein kinase, 2′-5′ oligoadenylate synthetases, ribonuclease L, Mx-GTPase, and ISG15.4, 5, 6
The ISG12 family is a group of small proteins (10–20 kDa) that contain a hydrophobic region (80–120 amino acids) referred to as the ISG12 motif.7, 8, 9 In silico analysis has identified four ISG12 genes (6–16, ISG12a, ISG12b, and ISG12c) in humans and three (ISG12a, ISG12b1, and ISG12b2) in mice.9 These genes have been poorly characterized and their physiological functions remain elusive. The expression of mouse ISG12b1 has been shown to significantly delay Sindbis virus-induced death in neonatal mice, providing evidence that ISG12b1 has a role in host antiviral defence.10 Additionally, mouse ISG12b1 is predominantly expressed in adipose tissue, specifically in the adipocytes,11, 12 wherein it hinders mitochondrial biogenesis and inhibits adipocyte differentiation.11 Human ISG12a is a mitochondrial protein that enhances sensitivity to DNA damage-induced apoptosis via perturbation of normal mitochondrial function.13 In contrast to human ISG12a, human 6–16, also known as G1P3, is a pro-survival protein. G1P3 blocks mitochondria-mediated apoptosis by inhibiting depolarization of the mitochondrial membrane potential (Δψm) in gastric cancer cells.14 G1P3 also antagonizes tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by inhibiting the intrinsic apoptotic pathway through mitochondrial stabilization in human myeloma cells.15 Altogether, these studies indicate that the ISG12 family proteins have diverse biological roles, depending on the cell/tissue types or microenvironments they occupy and the physiological responses they mediate.
Using microarray technology, we found that the expression of ISG12b2 was substantially increased in liver tissues from dengue virus (DENV)-infected mice compared with the control mice (Lu and Liao, unpublished data). In this study, we performed biochemical and functional analyses and showed that ISG12b2 localizes to the inner mitochondrial membrane (IMM) and mediates intrinsic apoptosis during viral infection. Our characterization of a new member of the ISG12 family provides additional understanding of the antiviral effects exerted by the ISG12 family.
Results
ISG12b2 is substantially induced both in vivo and in vitro in response to DENV
We performed real-time quantitative PCR and confirmed our microarray result that the level of ISG12b2 mRNA in liver tissues from DENV-infected mice was significantly increased as compared with that of the uninfected mice (Figure 1a). ISG12b2 expression was also significantly induced in a mouse hepatoma cell line, Hepa 1-6, stimulated with type I IFNs or poly(I:C) or infected with DENV (Figure 1b). Furthermore, induction of ISG12b2 by DENV infection is IFN dependent because mice deficient in STAT1, a transcription factor essential for type I IFN signalling, failed to induce ISG12b2 on DENV infection (Figure 1c).
ISG12b2 induction in response to DENV infection, IFNs, or poly(I:C). (a) ISG12b2 mRNA expression in livers from mock-infected (mock; n=4) or DENV-infected (n=5) mice was analysed by real-time quantitative PCR at 24 h post-infection. (b) ISG12b2 mRNA expression in Hepa 1-6 cells treated for 24 h with poly(I:C) (0.8 μg/ml), IFN-α (1000 U/ml), IFN-β (500 U/ml), or DENV (MOI=5) was analysed by real-time quantitative PCR. None, no treatment. (c) ISG12b2 mRNA expression in livers from mock-infected or DENV-infected WT or STAT1−/− mice (C57BL/6) was analysed by quantitative real-time PCR at 24 h post-infection. Data reflect the mean±S.E.M.; **P<0.01; ***P<0.001
Unprocessed and processed forms of ISG12b2 are generated in transfected cells
ISG12b2 encodes a putative transmembrane protein of 283 amino acids with two ISG12 motifs located at residues 8–90 and residues 133–215 (Figure 2a). The regions between the ISG12 motifs and at the C-terminal tail contain unique QXX (X represents any amino acid residue) repeats (Figure 2a). We raised a rabbit polyclonal antibody against ISG12b2, and generated full-length ISG12b2 (no tag) as well as N-terminal HA-tagged ISG12b2 (HA-ISG12b2) and C-terminal HA-tagged ISG12b2 (ISG12b2-HA) expression constructs. Western blot analysis of lysates from cells transfected with these ISG12b2 constructs using the anti-ISG12b2 antibody showed two distinct ISG12b2 species (Figure 2b, left panel). The two species of ISG12b2-HA migrated more slowly than those of HA-ISG12b2 or untagged ISG12b2. Notably, the anti-HA antibody detected the two polypeptide species in cells expressing ISG12b2-HA but not in cells expressing HA-ISG12b2 (Figure 2b, right panel). These results suggested that ISG12b2 contains a cleavable signal peptide at its N-terminus. Given that both polypeptides were confirmed by mass spectrometry to be authentic ISG12b2 species (data not shown) and that the MitoProt II program (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html) predicted a putative mitochondrial targeting sequence (MTS), which may be cleaved at residue 91 of ISG12b2,16, 17 we hypothesized that the low-molecular-weight species was generated because of the cleavage of ISG12b2 at residue 91 by a mitochondrial processing peptidase (MPP) and the high-molecular-weight species is the uncleaved ISG12b2. To further demonstrate that ISG12b2 contains a MTS, we constructed a plasmid encoding the N-terminal 93 amino acids of ISG12b2 fused to GFP (93-GFP) and examined transfected cell lysates by western blotting using an anti-GFP antibody. Two GFP-containing polypeptides were observed: uncleaved 93-GFP and GFP (Figure 2c, lower left panel). Additionally, the ISG12b2 mutant 94-283 with truncation of the first 93 residues including the putative MTS exhibited a single band with a molecular weight similar to that of the full-length ISG12b2 cleavage product (Figure 2c, lower right panel). Taken together, these results suggested that ISG12b2 contains a MTS that guides cleavage likely at residue 91 by MPP to generate a processed form.
Unprocessed and processed forms of ISG12b2 are generated in transfected cells. (a) Primary amino acid sequence of ISG12b2. Underlining, transmembrane helix; blue shading, ISG12 motifs; red lettering, QXX repeats. (b) Hepa 1-6 cells were transfected with empty vector or with plasmid encoding full-length untagged (no tag), N-terminal HA-tagged (HA-ISG12b2) or C-terminal HA-tagged (ISG12b2-HA) ISG12b2 and analysed by immunoblotting using anti-ISG12b2 or anti-HA. Unprocessed ISG12b2, arrows; processed ISG12b2, arrowheads. (c) Upper panel shows schematic diagram of full-length ISG12b2 (ISG12b2-GFP, FL-GFP; ISG12b2-HA, FL-HA), truncation mutant 93-GFP and 94-283-HA constructs. Lower left panel shows western blot analysis of lysates from Hepa 1-6 cells transfected with pEGFP-N1, full-length ISG12b2-GFP, or 93-GFP constructs using anti-GFP. Lower right panel shows western blot analysis of lysates from Hepa 1-6 cells transfected with empty vector, ISG12b2-HA, or truncated mutant 94-283-HA constructs using anti-ISG12b2. ISG, ISG12 motif
ISG12b2 is localized to the IMM
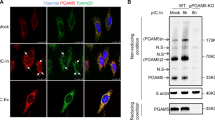
As ISG12b2 contains a MTS, it is likely localized to mitochondria. To test this possibility, we fractionated vesicular organelles from ISG12b2-HA-transfected HEK293T cells using 2.5–30% iodixanol gradient centrifugation. As expected, ISG12b2-HA was detected mainly in the mitochondria/ER fractions (Figure 3a). Immunostaining analysis by confocal microscopy also revealed the colocalization of ISG12b2 with DsRed2-Mito, a mitochondrial targeting fusion protein (Figure 3b). We then determined submitochondrial compartment localization of ISG12b2 by trypsin digestion of mitochondria isolated from transfected cells. Similar to the IMM-associated protein (the large subunit of complex II) and mitochondrial matrix protein (Hsp60), the processed form of ISG12b2 and a portion of the unprocessed form of ISG12b2 were sensitive to exogenously added trypsin only in the presence of Triton X-100 (Figure 3c). In contrast, the outer mitochondrial membrane (OMM) protein mitofusin 1 (Mfn1) was sensitive to trypsin digestion in the absence of Triton X-100 (Figure 3c). To further verify that ISG12b2 is an integral membrane protein, mitochondria were isolated from ISG12b2-HA-transfected Hepa 1-6 and resuspended in the cytosol extraction buffer or in a buffer containing 0.1 M Na2CO3 (pH 11.5), which releases the soluble and peripheral membrane-associated proteins into the supernatant. Western blotting showed that Hsp60 and complex II were released into the supernatant (Figure 3d). Like the integral membrane protein Mfn1, the majority of ISG12b2 was retained in the mitochondrial membrane pellet, although a portion of the processed form of ISG12b2 was released into the supernatant. Additional immmunogold electron microscopy also demonstrated the submitochondrial localization of ISG12b2 to the IMM (Figure 3e). Altogether, these results indicate that ISG12b2 is a transmembrane protein that localizes to the IMM.
ISG12b2 is localized to mitochondria. (a) Post-nuclear homogenates (PNH) from ISG12b2-HA-transfected HEK293T cells were fractionated by 2.5%–30% iodixanol gradient ultracentrifugation. Fractions were subjected to 12% SDS-PAGE followed by western blotting with anti-HA or antibodies specific for subcellular proteins including EEA1 (endosome), GM130 (Golgi), cytochrome c (mitochondria), or calnexin (ER). (b) NIH/3T3 cells co-transfected with the ISG12b2-HA and pDsRed2-Mito (red) constructs were immunostained with anti-HA (green) and counterstained with DAPI (blue). Images were obtained by confocal microscopy. Scale bar=5 μm. (c) Mitochondria isolated from Hepa 1-6 cells transfected with the ISG12b2-HA construct were subjected to trypsin digestion (50 μg/ml) in the presence or absence of 1% Triton X-100 at 37 °C for 1 h. Samples were subjected to 12% SDS-PAGE and subsequently immunoblotted with anti-ISG12b2, anti-Mfn1 (OMM protein), anti-Complex II large subunit (IMM-associated protein), or anti-Hsp60 (mitochondrial matrix protein). (d) Mitochondria isolated as described in c were incubated with or without 0.1 M Na2CO3 (pH 11.5) and centrifuged. The resultant supernatant (S) and pellet (P) were subjected to immunoblotting using anti-ISG12b2, anti-Mfn1, anti-complex II large subunit, or anti-Hsp60. (e) Hepa 1-6 cells transfected with ISG12b2-HA for 24 h were fixed, ultrathin sectioned, immunogold stained for HA, and visualized by transmission electron microscopy. Scale bar=500 nm
Overexpression of ISG12b2 leads to caspase-dependent cell death
Given that mitochondria execute apoptosis during viral infection and that ISG12b2 localizes to the mitochondria, we investigated whether ISG12b2 mediates apoptotic cell death. Hepa 1-6 cells transiently transfected with the ISG12b2 expression plasmid became rounded and retracted from the neighbouring cells, suggesting that overexpression of ISG12b2-induced apoptosis. To further identify the early-stage apoptosis and the late-stage cell death, we performed Annexin V and propidium iodide (PI) staining in the transfected cells. Flow cytometry analysis indicated that a significant population of cells expressing ISG12b2-GFP, but not GFP, was Annexin V positive (Figure 4a) and that the population of both Annexin V and PI-positive apoptotic cells increased with time (Figure 4b). Cells expressing ISG12b2-GFP also showed markedly increased in the population with sub-G1 DNA content as compared with cells expressing GFP (Figure 4c). Collectively, these data suggest that ISG12b2 mediates apoptotic cell death.
Expression of ISG12b2 leads to the release of cytochrome c, the impairment of Δψm and the apoptotic cell death. (a) Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP construct were subjected to Annexin V staining followed by flow cytometry analysis at 24 h post-transfection. (b) Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP construct were stained with Annexin V and PI at the indicated times and examined by flow cytometry. Cells positive for both Annexin V and PI were considered apoptotic. (c) Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP construct were analysed for sub-G1 DNA content by flow cytometry at 24 h post-transfection. (d) Hepa 1-6 cells were transfected with pEGFP-C1, ISG12b2-GFP, or ISG12b2-HA constructs in the presence of 40 μM Z-VAD-fmk and then were subjected to immunostaining using antibodies against cytochrome c (red) or HA (green) at 24 h post-transfection. Some ISG12b2-GFP- and ISG12b2-HA-expressing cells had no punctate cytochrome c signal (arrows), whereas some had low levels of the punctate cytochrome c signal (arrowheads). Asterisks indicate normal punctate cytochrome c signal in GFP-, ISG12b2-GFP-, and ISG12b2-HA-expressing cells. The percentage of cells with a normal punctate cytochrome c signal was quantified by confocal microscopy (6–11 different fields per sample), as shown in the bar graph at right. (e) Whole-cell lysates (WCL) and mitochondrial fractions (Mito) obtained from GFP- or ISG12b2-GFP-expressing cells were subjected to western blotting using anti-cytochrome c, anti-GFP, and anti-complex II large subunit. Asterisks indicate the presumably degraded form of GFP. The level of cytochrome c was quantified and normalized to complex II by densitometric analysis shown as bar graphs (right panel). (f) Δψm of Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP constructs was determined using MitoTracker Red staining followed by flow cytometry analysis at 24 h post-transfection. Cells with decreased MitoTracker Red signals were gated for analysis. Data represent the mean±S.E.M. and reflect one representative of three independent experiments
We next examined the effect of ISG12b2 on cytochrome c release from mitochondria, a key event in the process of intrinsic apoptosis. Immunostaining of cells transfected with the GFP expression vector showed a punctate cytochrome c pattern. However, ISG12b2-GFP- and ISG12b2-HA-expressing cells either completely lost (arrows) or had low levels (arrowheads) of punctate cytochrome c staining (Figure 4d). In addition, western blotting showed a significant decrease in cytochrome c in the mitochondrial fraction of ISG12b2-GFP-expressing cells as compared with GFP transfectants (Figure 4e), indicating that overexpression of ISG12b2 resulted in the release of cytochrome c from mitochondria. The amount of the large subunit of complex II was not affected by the overexpression of ISG12b2 (Figure 4e). As the release of cytochrome c from mitochondria is associated with the depolarization of Δψm during apoptosis, we next determined Δψm using MitoTracker Red CMXRos, which stains mitochondria in a Δψm-dependent manner. A sub-population of ISG12b2-GFP-expressing cells exhibited reduced MitoTracker Red signals (Figure 4f), suggesting that overexpression of ISG12b2 impaired Δψm, subsequently triggering mitochondria-mediated apoptosis.
To better understand the role of ISG12b2 in mitochondria-mediated apoptosis, we analysed caspase activation by western blot analysis. A substantially increase in the active (cleaved) form of caspase-3 (Cl-casp-3) and the cleaved form of poly (ADP-ribose) polymerase (Cl-PARP) was observed in ISG12b2-HA transfectants as compared with empty-vector transfectants (Figure 5a). In line with these results, fluorometric assays indicated that caspase-3, -9, and -8 activities were all significantly increased in ISG12b2-HA-expressing cells as compared with controls (Figure 5b). The activities peaked at 12 h post-transfection and declined at 24 h post-transfection (Figure 5b), which is likely due to the outgrowth of non-transfected cells over ISG12b2-transfected cells. Additionally, apoptosis induced by ISG12b2 overexpression was inhibited by Z-VAD-fmk, a pan-caspase inhibitor (Figure 5c). As caspase-9 and caspase-8 are the apical caspases in the intrinsic and extrinsic apoptotic pathways, respectively, we examined whether caspase-9 or caspase-8 was responsible for the activation of the downstream effector caspase-3. Caspase-3 was fully processed to yield the active form in ISG12b2-GFP-expressing cells (Figure 5d, lane 2).18 The generation of active form of caspase-3 was inhibited significantly by Z-VAD-fmk (Figure 5d, lane 6), moderately by Z-DEVD-fmk (a caspase-3 inhibitor; Figure 5d, lane 3) and Z-LEHD-fmk (a caspase-9 inhibitor; Figure 5d, lane 4), but not by Z-IETD-fmk (a caspase-8 inhibitor; Figure 5d, lane 5). Consistent with the data from caspase-3 activation, similar inhibitions on PARP cleavage were also observed (Figure 5d). Together, these results showed that ISG12b2 has a role in mitochondria-mediated apoptosis.
Overexpression of ISG12b2 induces caspase-dependent cell death. (a) The levels of cleaved caspase-3 (Cl-casp-3) and cleaved PARP (Cl-PARP) from Hepa 1-6 cells transfected for the indicated times with empty vector or ISG12b2-HA construct were analysed by western blotting using antibodies against the proteins indicated at left. Tubulin was analysed as a loading control. (b) Caspase-3, -9, and -8 activities in cells transfected as in a were quantified at 6, 12, and 24 h post-transfection by fluorometric assay. ISG12b2-HA expression in the transfectants was analysed by western blot (right). Actin was analysed as a loading control. Data represent the mean±S.E.M. and reflect one representative of three independent experiments. *P<0.05; **P<0.01; and ***P<0.001 versus the empty vector; ns, not significant. (c) The percentage of apoptosis (positive for both Annexin V and PI staining) in Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP construct in the presence of DMSO or 50 μM Z-VAD-fmk was measured at 9, 12, and 24 h post-transfection. Data represent the mean±S.E.M. and reflect one representative of three independent experiments. ***P<0.001 versus DMSO treatment groups. (d) Levels of Cl-casp-3 and Cl-PARP from Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP construct for 9 h in the presence of DMSO, Z-DEVD-fmk, Z-LEHD-fmk, Z-IETD-fmk, or Z-VAD-fmk were analysed by western blotting using antibodies against the proteins indicated at left. Actin was analysed as a loading control. The level of Cl-casp-3 in the western blot was quantified and normalized to the level of ISG12b2-GFP by densitometric analysis shown as bar graph (right panel). Data represent the mean±S.E.M.
Knockdown of ISG12b2 reduces DENV- or poly(I:C)-induced caspase-3 activation
As the induction of apoptosis reflects a mechanism of host defence against viral infection,19 we examined the role of ISG12b2 in apoptosis induced by DENV infection or by the viral genome analogue poly(I:C). We used siRNA to knockdown endogenous ISG12b2 and evaluated the cleaved forms of caspase-3, caspase-9, and PARP following DENV infection or poly(I:C) stimulation. Western blot analysis indicated that ISG12b2-specific siRNA completely abolished ectopic ISG12b2 expression in Hepa 1-6 cells (Figure 6a). As our antibody against ISG12b2 could not detect endogenous ISG12b2 in DENV-infected Hepa 1-6 cells, siRNA knockdown of endogenous ISG12b2 was evaluated by real-time quantitative PCR (Figures 6b and d, right panels). We found that the cleaved forms of caspse-3, caspase-9, and PARP were significantly decreased in ISG12b2 siRNA transfectants as compared with control siRNA transfectants following DENV infection (Figure 6b, left panel) or poly(I:C) stimulation (Figure 6d, left panel). Consistent with reduced caspase activation, the percentage of Annexin V-positive cells was also significantly decreased in ISG12b2 knockdown cells following DENV infection (Figure 6c). These data indicate that ISG12b2 contributes to apoptotic cell death in response to DENV or the viral mimetic dsRNA (poly(I:C)).
Knockdown of ISG12b2 reduces DENV- or poly(I:C)-induced caspse-3 activation. (a) Hepa 1-6 cells were co-transfected with ISG12b2-HA expression plasmid and non-targeting control siRNA (Ctrl) or targeting ISG12b2 siRNA (ISG) followed by western blot analysis using anti-ISG12b2. Actin was analysed as a loading control. (b) Hepa 1-6 cells transfected with non-targeting control siRNA (Ctrl) or targeting ISG12b2 siRNA (ISG) were infected with DENV at a MOI of 5 at 6 h post-transfection. Cells were then lysed using 1% Triton lysis buffer for protein detection or Trizol reagent for RNA extraction at 72 h after DENV infection. Western blot analysis using antibodies against indicated proteins in whole-cell lysates is shown in left panel. Actin was analysed as a loading control. Knockdown of endogenous ISG12b2 by siRNA was measured by real-time quantitative PCR shown in right panel. (c) Hepa 1-6 cells transfected with non-targeting control siRNA (Ctrl) or targeting ISG12b2 siRNA (ISG) were infected with DENV at a MOI of 5 and then subjected to Annexin V-Cy5 staining at 66 h post-infection. Data represent the mean±S.E.M. and reflect one representative of three independent experiments. *P<0.05. (d) Hepa 1-6 cells transfected with non-targeting control siRNA (Ctrl) or targeting ISG12b2 siRNA (ISG) were treated with 0.4 μg/ml poly(I:C) at 48 h post-transfection. Cells were then lysed using 1% Triton lysis buffer for protein detection or Trizol reagent for RNA extraction at 24 h after poly(I:C) treatment. Western blot analysis and real-time quantitative PCR were performed as the same as b
ISG12b2 is involved in both Bax/Bak- and mPT-mediated apoptosis
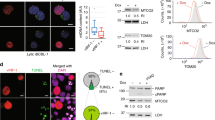
During apoptosis, the IMM protein adenine nucleotide translocase (ANT) physically interacts with the proapoptotic protein Bax to form pores, leading to increased mitochondrial membrane permeability and cell death.20, 21 We thus investigated whether ISG12b2, which is also an IMM protein, mediates cell death through interaction with Bax. Lysates of ISG12b2-HA-transfected Hepa 1-6 were immunoprecipitated for ISG12b2 using anti-HA agarose or for the active form of Bax using an active conformation specific antibody (6A7). Western blotting showed that Bax was associated with the ISG12b2-HA immunoprecipitates (Figure 7a, left panel), and ISG12b2-HA was present in the Bax immunoprecipitates (Figure 7a, right panel). Moreover, Bcl-2 family proapoptotic members-Bak and Bim, but not antiapoptotic members-Bcl-xL and BCL2L1, were found in the ISG12b2-HA immunoprecipitates (Figure7b and data not shown). The fact that ISG12b2 is associated with proapoptotic members of Bcl-2 family suggests ISG12b2 may mediate apoptosis via the Bax/Bak-dependent pathway. To determine whether Bax or Bak is essential for ISG12b2-mediated apoptosis, we examined the cleaved forms of both caspase-3 and PARP in wild-type (WT), Bax knockout (Bax−/−), Bak knockout (Bak−/−), and Bax-Bak double knockout (DKO) mouse embryonic fibroblasts (MEFs). Surprisingly, we found that both caspase-3 and PARP were processed as efficiently in ISG12b2-GFP-transfected DKO MEFs as in ISG12b2-GFP-transfected Bak−/− and WT MEFs (Figure 7c). Transfection per se induced caspase-3 and PARP processing in Bax−/− MEFs, likely because of their susceptibility to Lipofectamine 2000-induced toxicity (Figure 7c). We also observed that ISG12b2-GFP mediated the loss of Δψm and the release of cytochrome c in both WT and DKO MEFs (Figures 7d and e). These data indicate that ISG12b2 is able to mediate apoptosis via Bax/Bak-independent pathways.
ISG12b2 associates with Bax, Bak, and ANT and mediates both Bax/Bak-dependent and -independent apoptosis. (a) Hepa 1-6 cells transiently transfected with empty vector or ISG12b2-HA construct were lysed in 1% Triton lysis buffer followed by immunoprecipitation with anti-HA agarose (left panel) or lysed in 1% CHAPS lysis buffer followed by immunoprecipitation with anti-Bax (6A7) plus protein G beads (right panel). Immunoprecipitates were subjected to western blot analysis using anti-ISG12b2 or anti-Bax. As controls, whole-cell lysates (WCL) were subjected to western blot analysis (lower panels) using anti-ISG12b2 or anti-Bax. (b) Lysates from ISG12b2-HA-transfected Hepa 1-6 cells were immunoprecipitated with anti-HA agarose followed by western blot analysis using anti-Bak, anti-ANT, or anti-ISG12b2 (left panel). Lysates from ISG12b2-HA-transfected Hepa 1-6 cells were immunoprecipitated with anti-Bak or anti-ANT followed by western blot analysis using anti-HA (right panel). As controls, WCL were subjected to western blot analysis using the indicated antibodies. (c) Lysates from WT, Bax−/−, Bak−/−, and DKO MEFs transfected with GFP or ISG12b2-GFP construct for 28 h were analysed by western blotting using antibodies against the proteins indicated at left. Actin was analysed as a loading control. (d and e) WT and DKO MEFs transfected with ISG12b2-GFP expression construct in the presence of DMSO or 5 μM CsA for 28 h were subjected to MitoTracker Red staining (d) or to intracellular staining for cytochrome c (e). Cells positive for ISG12b2-GFP were gated to examine MitoTracker Red or cytochrome c signal by flow cytometry. **P<0.01. Data represent the mean±S.E.M. and reflect one representative of three independent experiments
Two major mechanisms have been proposed for the release of cytochrome c from mitochondria. One is mediated by Bax and Bak proteins, which induce mitochondrial outer membrane permeabilization by forming the oligomeric complex on OMM, known as the mitochondrial apoptosis-induced channel (MAC).22, 23, 24 The other is mediated by mitochondrial permeability transition (mPT) resulting from the opening of mPT pore, which is regulated by some stimuli or by the interaction with proapoptotic Bcl-2 family members.25, 26 Given that ISG12b2 is able to mediate apoptosis in the absence of Bax/Bak, and that ANT is a component of mPT pore in the IMM, we investigated whether ISG12b2 is associated with ANT and involved in mPT-mediated cytochrome c release. Western blot analysis showed that ANT was associated with ISG12b2-HA immunoprecipitates; reciprocally, ISG12b2-HA was present in the ANT immunoprecipitates (Figure 7b). Cyclosporine A (CsA), which binds to an activator of mPT pore component cyclophilin D, inhibits both the formation of mPT pore and the loss of Δψm, but does not prevent MAC-induced cytochrome c release.27, 28 As expected, CsA significantly inhibited the loss of Δψm in both ISG12b2-GFP-transfected WT and DKO MEFs (Figure 7d). However, CsA inhibited cytochrome c release only in ISG12b2-GFP-transfected DKO MEFs, but not in ISG12b2-GFP-transfected WT MEFs (Figure 7e). Collectively, these data indicated that ISG12b2 mediates cytochrome c release in WT MEFs as well as in Hepa 1-6 cells (Supplementary Figure 1) via Bax/Bak-dependent and/or mPT-independent pathways, however, in the absence of Bax/Bak, ISG12b2 mediates cytochrome c release in part via mPT-dependent (CsA-sensitive) pathways.
ISG12b2 forms oligomers in vivo
Bioinformatics analysis using the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM-2.0/) indicated that ISG12b2 contains seven glycine zipper motifs (one GXXXGXXXG and six GXXXGXXXA) and six putative transmembrane helices. The glycine zipper motifs are important for transmembrane helix interactions and may be involved in channel function.29 In addition, we found that ISG12b2 is associated with the aforementioned pore-forming proteins, Bax, Bak, and ANT (Figures 7a and b). We thus examined whether ISG12b2 forms oligomers. To this end, Hepa 1-6 were co-transfected with ISG12b2-HA and ISG12b2-GFP expression constructs followed by immunoprecipitation with anti-HA or anti-GFP. Western blot analysis using anti-ISG12b2 showed that ISG12b2-GFP was associated with the ISG12b2-HA immunoprecipitates, and reciprocally ISG12b2-HA was associated with the ISG12b2-GFP immunoprecipitates (Figure 8a), suggesting that ISG12b2 forms oligomers (Figure 8a). Our antibody failed to detect the endogenous ISG12b2 even in DENV-infected Hepa 1-6. It is likely due to either the low affinity of antibody-antigen complex or the low expression level of ISG12b2. As intestinal tissues express enormously high level of ISG12b2 mRNA (Supplementary Figure 2), our antibody may detect endogenous ISG12b2 in intestinal tissues. We therefore isolated mitochondria from small intestine tissues, incubated them with a membrane permeable crosslinker-DSS and analysed ISG12b2 by western blotting. Both unprocessed and processed forms of ISG12b2 were clearly detected in mitochondria of small intestinal tissues (Figure 8b). Importantly, a dimer form of the processed ISG12b2 was indeed observed in the presence of crosslinker-DSS, but the higher-order oligomers of ISG12b2 were difficult to see because of the high background (Figure 8b).
ISG12b2 forms oligomers. (a) Hepa 1-6 cells co-transfected with ISG12b2-HA and ISG12b2-GFP expression plasmids were lysed in 1% Triton X-100 lysis buffer followed by immunoprecipitation with anti-HA agarose or anti-GFP plus protein G beads. Co-immunoprecipitation of ISG12b2-HA and ISG12b2-GFP was detected by western blotting using anti-ISG12b2. The polypeptide migrating immediately below the processed form of ISG12b2-GFP likely results from the degradation of GFP portion in ISG12b2-GFP. Arrowheads, ISG12b2-HA; arrows, ISG12b2-GFP; *heavy chain. (b) Mitochondria isolated from the small intestinal tissues were incubated with DSS at the indicated concentrations for 30 min, lysed, and subjected to western blot analysis using anti-ISG12b2
Discussion
In this study, we characterized the mouse ISG12b2, a novel IFN-induced IMM protein that is involved in the regulation of intrinsic apoptotic cell death. Overexpression of ISG12b2 leads to induction of mitochondria-mediated apoptosis, whereas knockdown of ISG12b2 followed by DENV infection reduces apoptosis. Of particular interest, ISG12b2 forms oligomers and is associated with proteins involved in pore-forming complexes including Bax, Bak, and ANT, suggesting that ISG12b2 likely cooperates with those pore-forming proteins to form pores and/or channels and thus mediates apoptosis. However, the underlying mechanisms of ISG12b2 action remain to be investigated.
The first line of host defence against viral infection is mainly mediated by the type I IFN system, which generates an extremely powerful antiviral response.30, 31 On viral infection, the host germline-encoded pattern recognition receptors, such as TLR3,32 TLR7,33, 34 TLR8,35, 36 RIG-I,37 and MDA538 recognize the virus or viral products and induce type I IFNs, such as IFN-α/β. Type I IFNs then bind to their cognate receptors and trigger transcription of a diverse set of genes referred to as ISGs, many of which exhibit direct or indirect antiviral function.30 Among ISGs, the biochemical and physiological functions of the ISG12 family are largely unknown. Two ISG12 members have opposing influences on apoptotic cell death. Human ISG12a mediates pro-apoptotic effects induced by DNA damage,13 whereas human 6–16 (G1P3) antagonizes TRAIL-mediated apoptosis in cancerous cell lines.15 This study has demonstrated that mouse ISG12b2 triggers mitochondria-mediated apoptosis during viral infection. Although these three ISG12 proteins regulate apoptosis, their mechanisms of action differ. Human ISG12a regulates intrinsic apoptosis, which seems to enhance sensitivity to DNA damage-induced apoptosis;13 human 6–16 regulates extrinsic apoptosis;15 mouse ISG12b2 initiates intrinsic apoptosis on virus infection or type I IFN stimulation. Of note, a previous study showed that ISG12b1 protects neonatal mice from lethal encephalitis induced by Sindbis virus infection, but no mechanism was demonstrated.10 This study demonstrated that mouse ISG12b2 is dramatically induced by DENV infection both in vivo and in vitro. Given the high similarity of amino acid sequences between mouse ISG12b2 and mouse ISG12b1, mouse ISG12b1 may trigger apoptosis in defence against Sindbis virus infection in a manner analogous to that observed for mouse ISG12b2 following DENV infection.
We have demonstrated that ISG12b2 not only forms oligomers but also associates with proteins involving in pore formation induced by apoptosis, such as Bax, Bak, and ANT, in ISG12b2-transfected Hepa 1-6. Given the oligomerization of ISG12b2 and the association of ISG12b2 with Bax/Bak, it is plausible that ISG12b2 forms pores in the IMM and cooperates with the pores formed by Bax and Bak in the OMM to create a conduit at the inner-outer membrane contact site, dismantling mitochondria, subsequently resulting in apoptosis. ANT, a component of the mPT pore, has been reported to form nonspecific pores in the IMM and directly interact with Bax to increase mitochondrial membrane permeability, subsequently triggering cell death.20, 21, 25 Our findings on the association of ISG12b2 with both ANT and Bax prompted us to propose that ISG12b2 may form a pore complex of ISG12b2-ANT-Bax to facilitate mitochondria-mediated apoptosis.
Cytochrome c release and apoptosis can take place independent of Bax/Bak, but dependent on mPT.39, 40 Indeed, we observed that ISG12b2-mediated cytochrome c release in DKO MEFs was inhibited by CsA, a mPT inhibitor. These results suggest that ISG12b2 is likely involved in mPT-dependent apoptosis. As our finding showed the association of ISG12b2 with ANT, it is tempting to speculate that the effect of ISG12b2 on the mPT is related to the regulation of ANT. We cannot rule out the possibility that ISG12b2 activates other unidentified mitochondrial target(s) that participate in the regulation of mPT. Of note, we found that CsA did not completely inhibit the cytochrome c release, suggesting that ISG12b2 may be involved in an unidentified apoptotic pathway independent of both Bax/Bak and mPT. Altogether, our results suggest that ISG12b2 has a role in mediating mitochondria-mediated apoptosis via Bax/Bak-dependent or Bax/Bak-independent but mPT-dependent pathways.
We also defined some biochemical characteristics of ISG12b2. First, ISG12b2 has two forms – unprocessed and processed forms. The processed form is likely due to the cleavage of the unprocessed form by MPP. Second, the apparent molecular weights of both forms of ISG12b2 are ∼10 kDa greater than their predicted molecular weights (Figure 2). This cannot be explained by posttranslational modifications including sumoylation and ubiquitination (Supplementary Figure 3), although we cannot exclude some other unidentified posttranslational modification. Alternatively, the aberrant mass may be due to ‘gel shifting’, in which the hydropathy of transmembrane regions of membrane proteins alter SDS binding, affecting protein migration in SDS-PAGE.41
ISGs are critical molecules induced in viral infections and the study of ISGs is of importance for understanding their roles in antiviral infection. Our study has characterized a novel ISG member-mouse ISG12b2, unveiling the functional activity of one more member of the ISG family.
Materials and Methods
Mice
Mice (C57BL/6) were maintained under specific pathogen-free conditions at the Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan, and were used at 6–8 weeks old. All animal experiments were approved by the Institutional Animal Care and Utilization Committee at Academia Sinica and were performed in accordance with institutional guidelines. STAT1−/− mice were kindly provided by Dr. Chien-Kuo Lee (National Taiwan University, Taipei, Taiwan).
Cell lines and culture
The mouse hepatoma cell line Hepa 1-6 and the human kidney cell line HEK293T were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The mouse fibroblast NIH/3T3 cell line was purchased from the Bioresource Collection and Research Center (HsinChu, Taiwan). Immortalized MEFs derived from WT, Bax knockout (Bax−/−), Bak knockout (Bak−/−), or Bax-Bak DKO mice were generously provided by Dr. Craig Thompson (University of Pennsylvania, Philadelphia, PA, USA).42 These cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2 mM L-glutamine (Invitrogen, Carlsbad, CA, USA) at 37 °C under 5% CO2 in a humidified atmosphere.
Construction of plasmids
Reverse transcriptase PCR amplification of total RNA from livers of C57BL/6 mice was initially performed to obtain the full-length ISG12b2 cDNA, which was then amplified by PCR using different primer pairs to obtain the various tagged full-length and truncated forms of ISG12b2 for subsequent cloning into pcDNA3.1/Zeo (+) (Invitrogen) or pEGFP-N1 (Clontech, Mountain View, CA, USA). The forward and reverse primers used for amplification of various HA-tagged full-length or truncated ISG12b2 or GFP-fused full-length or truncated ISG12b2 are shown in Supplementary Table 1.
Plasmids and siRNA transfection
Cells were transfected with plasmids encoding various full-length and truncated ISG12b2 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfection of non-targeting control siRNA or targeting ISG12b2 siRNA (5′-CAUUGCAGCTGGGTCCATATT-3′) (Dharmacon, Lafayette, CO, USA) into Hepa 1-6 cells were also performed by Lipofectamine 2000.
Cell treatment
Hepa 1-6 cells were seeded in six-well plates at 6 × 105 cells per well. After overnight culture, cells were transfected with poly(I:C) (Sigma-Aldrich, St. Louis, MO, USA) using Lipofectamine 2000 (Invitrogen), treated with mouse IFN-αA and mouse IFN-β (PBL, Piscataway, NJ, USA), or infected with DENV at a multiplicity of infection (MOI) of 5. At 24 h post-treatment, cells were subjected to further analysis of ISG12b2 expression. For CsA experiment, cells pretreated with 5 μM CsA for 1 h at 37 °C were subjected to DNA transfection followed by analysis of Δψm and cytochrome c release.
ISG12b2 antibody generation
A peptide comprising residues 95 to 112 of ISG12b2 was synthesized and conjugated with ovalbumin, and the ovalbumin-conjugated peptide was used to raise polyclonal antibodies against the ISG12b2 in New Zealand rabbits (LTK BioLaboratories, Taoyuan, Taiwan).
DENV preparation and infection
DENV type 2 (New Guinea C strain) was prepared as described.43 For in vivo DENV infection, C57BL/6 mice were subjected to intravenous injection with 0.5 ml PBS or with DENV (3 × 107 plaque-forming unit) and simultaneously injected intracerebrally with 30 μl PBS to break blood–brain barrier. At 24 h post-treatment, mice were killed and the livers were perfused with 3 ml of PBS before isolation of total RNA. For in vitro DENV infection, Hepa 1-6 cells were infected with DENV at a MOI of 5 for 2 h, washed with DMEM once, and further cultured in DMEM containing 10% FBS for the indicated times.
Real-time quantitative PCR analysis
Total RNA was extracted from mouse livers or Hepa 1-6 cells by Trizol reagent (Invitrogen), and 3 μg total RNA was subjected to reverse transcription using an oligo(dT) primer and SuperScript III reverse transcriptase (Invitrogen). For real-time quantitative PCR amplification of cDNA, the 2–ΔΔCT method was used to quantify the relative changes in gene expression. Amplification of ISG12b2 and GAPDH cDNAs was performed using a TaqMan Gene Expression assay kit containing a mixture of unlabeled PCR primers and TaqMan FAM-labeled MGB probe specific for each gene (Applied Biosystems, Foster City, CA, USA). The PCR reaction conditions were as follows: 50 °C for 2 min; 95 °C for 10 min; 40 cycles at 95 °C for 15 s, and 60 °C for 60 s. ISG12b2-specific PCR products were quantified using an ABI 7500 Real-Time PCR System (Applied Biosystems) and normalized to the level of GAPDH gene expression.
Confocal microscopic analysis of immunofluorescence staining
For detection of ISG12b2 subcellular localization, NIH/3T3 cells (2 × 104) grown on coverslips in a 24-well plate were co-transfected with the ISG12b2-HA construct and pDsRed2-Mito (Clontech). At 24 h post-transfection, cells were washed and stained with rabbit anti-HA followed by anti-rabbit IgG F(ab’)2 conjugated with Alexa 488 (Molecular Probes, Eugene, OR, USA). For cytochrome c detection, Hepa 1-6 cells (1.2 × 105) grown on coverslips in a 24-well plate were transfected with pEGFP-C1, ISG12b2-GFP, or ISG12b2-HA constructs and cultured in the presence of 40 μM Z-VAD-fmk (BACHEM, Bubendorf, Switzerland). After 24 h, transfectants were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100/PBS for 5 min, washed three times with PBS, incubated in PBS containing 3% BSA for 1 h at room temperature, and immunostained with rabbit anti-HA plus mouse anti-cytochrome c (BD Pharmingen, San Jose, CA, USA) for 90 min at room temperature. After three washes with PBS, cells were further incubated with goat anti-rabbit IgG F(ab’)2 conjugated with Alexa 488 and goat anti-mouse IgG conjugated with Alexa 594 (Molecular Probes) for 30 min followed by counter staining with 1 μg/ml DAPI (Molecular Probes) for 5 min. Coverslips were mounted on microscope slides using fluorescence mounting medium (Dako Cytomation, Glostrup, Denmark) and subjected to confocal microscopy. Images were obtained using a Radiance 2100 (Bio-Rad, Hertfordshire, UK) or LSM510 (PerkinElmer, Waltham, MA, USA) laser scanning confocal microscope.
Subcellular fractionation
HEK293T cells transfected with the ISG12b2-HA expression construct were homogenized in homogenization buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4) containing protease inhibitors (10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM PMSF) using a Dounce homogenizer (Wheaton, Millville, NJ, USA) and centrifuged at 800 × g for 5 min. The resulting supernatants (post-nuclear homogenates) were applied to a 2.5–30% (v/v) linear iodixanol gradient (Axis-Shield, Oslo, Norway) followed by centrifugation by Beckman Coulter Optima L-90K ultracentrifuge (Beckman Coulter, Brea, CA, USA) for 3.5 h at 200 000 × g in a swinging-bucket rotor (SW41). After centrifugation, a series of 1 ml fractions were carefully collected from the top to the bottom of the sample, and the fractions were subjected to 12% SDS-PAGE followed by immunoblot analysis using anti-HA, anti-EEA1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-GM130 (BD Transduction Laboratories, San Jose, CA, USA), anti-cytochrome c (BD Pharmingen), or anti-calnexin (Affinity BioReagents, Rockford, IL, USA).
Immunogold electron microscopy
Hepa 1-6 cells transfected with ISG12b2-HA for 24 h were fixed for 1 h at room temperature with 4% paraformaldehyde and 0.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3) (CAB) and post-fixed with 4% paraformaldehyde, 0.1% glutaraldehyde, and 0.05% tannic acid in 0.1 M CAB. After washing with CAB, the cells were suspended in cryoprotectant (30% glycerol), cooled to 4 °C, immersed in liquid nitrogen, and subsequently transferred to freeze substitution. The cells were then embedded in HM20 resin (EMS, Hatfield, PA, USA) and ultrathin sectioned. The sections were blocked with 0.1% BSA/PBS for 1 h at room temperature, incubated with rabbit anti-HA for overnight at 4 °C, washed, and further incubated with goat anti-rabbit IgG conjugated with 18 nm gold (Jackson Lab, Bar Harbor, ME, USA) for 1 h at room temperature. The sections were then subjected to washing with PBS followed by fixation with 1% glutaraldehyde for 10 min at room temperature. After washing with distilled water, sections were dried, coated with Formvar (EMS), and stained with uranyl acetate and lead citrate. All sections were observed with a Hitachi H7500 electron microscope (Tokyo, Japan) at a 75-kV acceleration voltage.
Mitochondria isolation, trypsin digestion, and alkaline extraction
Mitochondrial and cytosolic fractions of Hepa 1-6 cells transfected with the ISG12b2-HA expression construct were isolated using a Mitochondria/Cytosol Fractionation Kit (BioVision, Mountain View, CA, USA). The isolated mitochondrial fraction (20 μg) was treated with 50 μg/ml trypsin (Promega, Madison, WI, USA) at 37 °C for 1 h in the presence or absence of 1% Triton X-100. The digested samples were subjected to 12% SDS-PAGE followed by western blotting with anti-ISG12b2, anti-Complex II large subunit (Molecular Probes), anti-Mfn1, or anti-Hsp60 (Santa Cruz Biotechnology). For alkaline extraction, the isolated mitochondrial pellets (∼55 μg) were resuspended in 100 μl cytosol extraction buffer or 0.1 M Na2CO3 (pH 11.5) and incubated on ice for 40 min followed by centrifugation at 20 000 × g for 1 h. The insoluble membrane fractions were then dissolved in Laemmli buffer and subjected to 12% SDS-PAGE followed by western blotting using the indicated antibodies.
Detection of apoptosis
For detection of apoptotic cells by Annexin V staining, Hepa 1-6 cells transfected with pEGFP-C1 or ISG12b2-GFP constructs were cultured in the absence or presence of 40 μM Z-VAD-fmk, harvested at the indicated times, and stained for 5 min at room temperature with Annexin V-Cy5 (BioVision) and 1 μg/ml PI (Sigma-Aldrich). Stained cells were analysed using a FACS Calibur flow cytometer and Cell Quest software (BD Biosciences, San Jose, CA, USA). For detection of sub-G1 DNA content, ∼2 × 106 transfected cells were harvested, fixed with 5 ml cold 70% ethanol, and incubated at −20 °C overnight. Fixed cells were centrifuged at 500 × g, washed once with 5 ml PBS, and resuspended in 0.25 ml PBS; 1 ml DNA extraction buffer (0.19 M Na2HPO4/4 mM citric acid, pH 7.8) was added, and cells were further incubated for 5 min at room temperature to facilitate extraction of low-molecular-weight DNA. After centrifugation at 700 × g for 10 min, cells were then stained with 20 μg/ml PI in the presence of 200 μg/ml RNase A in PBS for 30 min at room temperature followed by flow cytometry analysis. For detection of apoptosis by caspase activation assay, cells transfected with the indicated expression plasmid in the absence or presence of 50 μM Z-VAD-fmk, Z-DEVE-fmk, Z-LEHD-fmk, or Z-IETD-fmk (the latter three from R&D systems, Minneapolis, MN, USA) were lysed in 1% Triton lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM PMSF), and the resulting whole-cell lysates (30 μg) were subjected to12% SDS-PAGE followed by immunoblotting with anti-cleaved PARP (BD Pharmingen), anti-cleaved caspase-3 (Cell Signaling Technology, Danvers, MA, USA), anti-ISG12b2, anti-tubulin (Abcam, Cambridge, MA, USA), or anti-actin (Sigma-Aldrich). In some experiments, caspase activity was determined by fluorometry using a Caspase Fluorometric Assay Kit (BioVision). Briefly, whole-cell lysates (30–50 μg per sample) were incubated with the caspase substrates DEVD-AFC (caspse-3), LEHD-AFC (caspase-9), or IETD-AFC (caspase-8) at 37 °C for 1.5 h. After incubation, released AFC was measured using a fluorescence microplate reader (Device, Sunnyvale, CA, USA) at an excitation wavelength of 400 nm and an emission wavelength of 505 nm.
Detection of Δψm and cytochrome c release
For detection of Δψm, cells transfected with pEGFP-C1 or ISG12b2-GFP expression plasmids were trypsinized and stained with 150 nM MitoTracker Red CMXRos (Molecular Probes) in culture medium for 30 min at 37 °C followed by flow cytometry analysis; the GFP or ISG12b2-GFP-positive population was subjected for analysis of Δψm. For detection of cytochrome c release, mitochondrial and cytosolic fractions were isolated from transfected cells using a Mitochondria/Cytosol Fractionation Kit. Whole-cell lysates and mitochondrial fractions (10 μg) were subjected to 12% SDS-PAGE followed by immunoblotting with anti-cytochrome c, anti-GFP (Santa Cruz Biotechnology), or anti-complex II large subunit. For intracellular staining of cytochrome c, cells were trypsinized, centrifuged and then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After washing with FACS buffer (1% FBS, 10 mM HEPES, and 0.1% NaN3 in HBSS), cells were incubated in blocking buffer (5% non-fat milk, 0.1% saponin, 0.5% BSA, and 0.05% NaN3 in PBS) for 30 min at room temperature followed by incubation with mouse anti-cytochrome c for 30 min at room temperature. After twice washes, the cells were further incubated with goat anti-mouse IgG F(ab’)2 conjugated with Alexa 647 for 30 min at room temperature followed by two washes. The cells were then subjected to flow cytometry analysis; the GFP or ISG12b2-GFP-positive population was gated for analysis of cytochrome c release.
Immunoprecipitation and western blotting
Hepa 1-6 cells were transfected with ISG12b2-HA expression plasmid, lysed in 1% CHAPS lysis buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 1% CHAPS, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM PMSF) for immunoprecipitation of the active Bax or in 1% Triton lysis buffer for immunoprecipitation of other proteins, and centrifuged at 16 000 × g for 15 min. Supernatants (1 mg) were pre-cleared with protein G-agarose beads (Millipore, Billerica, MA, USA). Precleared lysates were incubated with 2 μg anti-Bax (6A7) (BD Biosciences), with 3 μg anti-ANT, or with 2 μg anti-Bak (Santa Cruz Biotechnology) at 4 °C overnight followed by the addition of 30 μl protein G-agarose beads and incubation for 1 h to precipitate the active Bax, ANT, or Bak proteins. The immunoprecipitates were subjected to 12% SDS-PAGE followed by western blotting using anti-ISG12b2. For immunoprecipitation of ISG12b2, pre-cleared lysates were incubated with anti-HA agarose (Sigma-Aldrich) for overnight at 4 °C, and the immunoprecipitates were subjected to 12% SDS-PAGE followed by western blotting with anti-Bax (N20), anti-ANT, anti-Bak, anti-HA, or anti-ISG12b2. To examine ISG12b2 self-association, lysates from Hepa 1-6 cells co-transfected with ISG12b2-HA and ISG12b2-GFP expression plasmids were incubated with anti-HA agarose or anti-GFP plus protein G-agarose beads, and the immunoprecipitates were subjected to 12% SDS-PAGE followed by western blotting with anti-ISG12b2.
Chemical crosslinking
The small intestinal tissues from C57BL/6 mice were excised, cut and minced into small pieces, washed with PBS twice, and then incubated in homogenization buffer (0.25 M sucrose, 10 mM KCl, 1 mM EDTA, 10 mM HEPES, pH 7.4). The tissues were homogenized using Dounce homogenizer, and centrifuged at 700 × g for 10 min at 4 °C to remove cell debris. The homogenates were further centrifuged at 10 000 × g for 15 min at 4 °C to obtain the mitochondrial pellets. The mitochondrial pellets were resuspended in PBS and the concentration was determined using Bradford protein assay (Bio-Rad). Thirty μg of mitochondria were incubated with 0, 0.5, 1.0, 2.0, or 5.0 mM DSS (Pierce, Rockford, IL, USA) at room temperature for 30 min followed by the addition of 10 mM Tris-HCl (pH 7.4) at room temperature for 15 min to terminate the reaction. The mitochondria were then centrifuged, dissolved in Laemmli buffer and subjected to 10% SDS-PAGE followed by western blotting with anti-ISG12b2.
Statistical analysis
The unpaired Student's t-test was applied to evaluate the differences between experimental groups. P≤0.05 was considered statistically significant.
Abbreviations
- IFN:
-
interferon
- ISG:
-
IFN-stimulated gene
- DENV:
-
dengue virus
- IMM:
-
inner mitochondrial membrane
- OMM:
-
outer mitochondrial membrane
- Δψm:
-
mitochondrial membrane potential
- MTS:
-
mitochondrial targeting sequence
- MPP:
-
mitochondrial processing peptidase
- PARP:
-
poly (ADP-ribose) polymerase
- ANT:
-
adenine nucleotide translocase
- mPT:
-
mitochondrial permeability transition
- CsA:
-
cyclosporine A
References
Isaacs A, Lindenmann J . Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 1957; 147: 258–267.
de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol 2001; 69: 912–920.
Der SD, Zhou A, Williams BR, Silverman RH . Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA 1998; 95: 15623–15628.
Malakhova OA, Zhang DE . ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem 2008; 283: 8783–8787.
Sadler AJ, Williams BR . Interferon-inducible antiviral effectors. Nat Rev Immunol 2008; 8: 559–568.
Silverman RH . Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 2007; 81: 12720–12729.
Gjermandsen IM, Justesen J, Martensen PM . The interferon-induced gene ISG12 is regulated by various cytokines as the gene 6-16 in human cell lines. Cytokine 2000; 12: 233–238.
Martensen PM, Justesen J . Small ISGs coming forward. J Interferon Cytokine Res 2004; 24: 1–19.
Parker N, Porter AC . Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12. BMC Genomics 2004; 5: 8.
Labrada L, Liang XH, Zheng W, Johnston C, Levine B . Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J Virol 2002; 76: 11688–11703.
Li B, Shin J, Lee K . Interferon-stimulated gene ISG12b1 inhibits adipogenic differentiation and mitochondrial biogenesis in 3T3-L1 cells. Endocrinology 2009; 150: 1217–1224.
Ohgaki S, Iida K, Yokoo T, Watanabe K, Kihara R, Suzuki H et al. Identification of ISG12b as a putative interferon-inducible adipocytokine which is highly expressed in white adipose tissue. J Atheroscler Thromb 2007; 14: 179–184.
Rosebeck S, Leaman DW . Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis 2008; 13: 562–572.
Tahara Jr E, Tahara H, Kanno M, Naka K, Takeda Y, Matsuzaki T et al. G1P3, an interferon inducible gene 6-16, is expressed in gastric cancers and inhibits mitochondrial-mediated apoptosis in gastric cancer cell line TMK-1 cell. Cancer Immunol Immunother 2005; 54: 729–740.
Cheriyath V, Glaser KB, Waring JF, Baz R, Hussein MA, Borden EC . G1P3, an IFN-induced survival factor, antagonizes TRAIL-induced apoptosis in human myeloma cells. J Clin Invest 2007; 117: 3107–3117.
Claros MG, Vincens P . Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 1996; 241: 779–786.
Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z et al. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 2001; 9: 615–625.
Inoue S, Browne G, Melino G, Cohen GM . Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ 2009; 16: 1053–1061.
Barber GN . Host defense, viruses and apoptosis. Cell Death Differ 2001; 8: 113–126.
Belzacq AS, Vieira HL, Verrier F, Vandecasteele G, Cohen I, Prevost MC et al. Bcl-2 and Bax modulate adenine nucleotide translocase activity. Cancer Res 2003; 63: 541–546.
Marzo I, Brenner C, Zamzami N, Jurgensmeier JM, Susin SA, Vieira HL et al. Bax and adenine nucleotide translocator cooperate in the mitochondrial control of apoptosis. Science 1998; 281: 2027–2031.
Nechushtan A, Smith CL, Lamensdorf I, Yoon SH, Youle RJ . Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J Cell Biol 2001; 153: 1265–1276.
Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I et al. Inhibition of Bax channel-forming activity by Bcl-2. Science 1997; 277: 370–372.
Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002; 111: 331–342.
Kroemer G, Galluzzi L, Brenner C . Mitochondrial membrane permeabilization in cell death. Physiol Rev 2007; 87: 99–163.
De Marchi U, Campello S, Szabo I, Tombola F, Martinou JC, Zoratti M . Bax does not directly participate in the Ca(2+)-induced permeability transition of isolated mitochondria. J Biol Chem 2004; 279: 37415–37422.
Martinez-Caballero S, Dejean LM, Kinnally KW . Some amphiphilic cations block the mitochondrial apoptosis-induced channel MAC. FEBS Lett 2004; 568: 35–38.
Eskes R, Antonsson B, Osen-Sand A, Montessuit S, Richter C, Sadoul R et al. Bax-induced cytochrome C release from mitochondria is independent of the permeability transition pore but highly dependent on Mg2+ ions. J Cell Biol 1998; 143: 217–224.
Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU . Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA 2005; 102: 14278–14283.
Basler CF, Garcia-Sastre A . Viruses and the type I interferon antiviral system: induction and evasion. Int Rev Immunol 2002; 21: 305–337.
Theofilopoulos AN, Baccala R, Beutler B, Kono DH . Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 2005; 23: 307–336.
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA . Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001; 413: 732–738.
Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C . Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004; 303: 1529–1531.
Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci USA 2004; 101: 5598–5603.
Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004; 303: 1526–1529.
Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H et al. Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat Immunol 2002; 3: 499.
Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5: 730–737.
Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K et al. Shared and unique functions of the DExD/H-box helicases RIG-I MDA5, and LGP2 in antiviral innate immunity. J Immunol 2005; 175: 2851–2858.
Mullauer FB, Kessler JH, Medema JP . Betulinic acid induces cytochrome c release and apoptosis in a Bax/Bak-independent, permeability transition pore dependent fashion. Apoptosis 2009; 14: 191–202.
Wan KF, Chan SL, Sukumaran SK, Lee MC, Yu VC . Chelerythrine induces apoptosis through a Bax/Bak-independent mitochondrial mechanism. J Biol Chem 2008; 283: 8423–8433.
Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM . Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci USA 2009; 106: 1760–1765.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 2001; 292: 727–730.
Hsieh MF, Lai SL, Chen JP, Sung JM, Lin YL, Wu-Hsieh BA et al. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J Immunol 2006; 177: 1855–1863.
Acknowledgements
We are grateful to Dr. Craig Thompson for providing MEFs derived from wild-type, Bax−/−, Bak−/−, Bax/Bak double knockout mice, to Dr. Steve S.L. Chen for providing antibodies to various subcellular organelles, to Dr. Chien-Kuo Lee for providing STAT1 knockout mice, to Dr. Hsiu-Ming Shih for providing FLAG-tagged ubiquitin and FLAG-tagged SUMO, to Mr. Hsin-Lin Lu for preparing dengue virus, to Dr. Pauline Yen for editing this paper, and to Core facilities for helping with confocal microscopy, flow cytometry, and transmission electron microscopy. This work was supported by grants from the National Science Council and Academia Sinica in Taiwan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by H-U Simon
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Lu, MY., Liao, F. Interferon-stimulated gene ISG12b2 is localized to the inner mitochondrial membrane and mediates virus-induced cell death. Cell Death Differ 18, 925–936 (2011). https://doi.org/10.1038/cdd.2010.160
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.160
Keywords
This article is cited by
-
Identification and validation of ferroptosis-related genes in patients infected with dengue virus: implication in the pathogenesis of DENV
Virus Genes (2023)
-
Transcriptome profiling of the rat retina after optic nerve transection
Scientific Reports (2016)
-
Oncolytic virotherapy
Nature Biotechnology (2012)