Abstract
Mutations in PARK7 DJ-1 have been associated with autosomal-recessive early-onset Parkinson's disease (PD). This gene encodes for an atypical peroxiredoxin-like peroxidase that may act as a regulator of transcription and a redox-dependent chaperone. Although large gene deletions have been associated with a loss-of-function phenotype, the pathogenic mechanism of several missense mutations is less clear. By performing a yeast two-hybrid screening from a human fetal brain library, we identified TRAF and TNF receptor-associated protein (TTRAP), an ubiquitin-binding domain-containing protein, as a novel DJ-1 interactor, which was able to bind the PD-associated mutations M26I and L166P more strongly than wild type. TTRAP protected neuroblastoma cells from apoptosis induced by proteasome impairment. In these conditions, endogenous TTRAP relocalized to a detergent-insoluble fraction and formed cytoplasmic aggresome-like structures. Interestingly, both DJ-1 mutants blocked the TTRAP protective activity unmasking a c-jun N-terminal kinase (JNK)- and p38-MAPK (mitogen-activated protein kinase)-mediated apoptosis. These results suggest an active role of DJ-1 missense mutants in the control of cell death and position TTRAP as a new player in the arena of neurodegeneration.
Similar content being viewed by others
Main
Parkinson's disease (PD) is the second most common progressive neurodegenerative disorder, affecting 1–2% of all individuals above the age of 65 years. Its neuropathological hallmark is the selective degeneration of subsets of mesencephalic dopaminergic cells and the formation of proteinaceous cytoplasmic aggregates called Lewy bodies.1
Several studies implicate the ubiquitin–proteasome system in PD pathogenesis.2 Synthetic proteasome inhibitors preferentially affect catecholaminergic neurons, leading to cell death. Furthermore, key ubiquitin–proteasome system elements are altered in PD post-mortem brains.3, 4, 5
The identification of genes (PARK1–14) associated with familial PD has provided crucial insights into the pathogenic mechanisms.1 The ubiquitin ligase parkin (PARK2) and the ubiquitin C-terminal hydrolase-L1 (UCH-L1) (PARK5) have been implicated in the ubiquitin–proteasome system function. Interestingly, upon treatment with proteasome inhibitors, they formed insoluble aggregates and were recruited to a juxtanuclear aggresome-like inclusion that resembled the Lewy body.6, 7
Autosomal-recessive early-onset PD has been associated with mutations in PARK7/DJ-1.8 Functional DJ-1 is a dimer that acts as an atypical peroxiredoxin-like peroxidase as well as a regulator of transcription and a chaperone.9, 10, 11, 12 Interestingly, ectopic DJ-1 expression protects cells from death induced by a variety of insults.13
DJ-1 loss in humans causes PD.14 DJ-1 knock-out (KO) mice and flies showed increased vulnerabilities to neurotoxic agents but no signs of dopaminergic cell death.15, 16, 17
PD families may also present missense mutations of DJ-1 in homozygous (L166P, M26I and E64D) and heterozygous forms (A104T and D149A).14 How these mutants abolish or modify DJ-1 functions is a matter of debate, as a common mechanism of action for the various DJ-1 mutants is still lacking. M26I and L166P are the most studied DJ-1 missense mutations. Although M26I dimer formation and protein levels reported in the literature may vary, differences with wild-type (WT) DJ-1 are small and of unclear biological significance. On the contrary, L166P is very unstable and its expression level lower than WT.18, 19, 20, 21
Together with a loss of function, DJ-1 missense mutations may be involved in pathological protein–protein interactions that may lead to a ‘gain-of-function’ behavior. The search for common protein partners that distinguish mutant DJ-1s from WT has been so far limited, with the important exception of an increased binding to parkin shared by M26I and L166P.22
In this paper, we aimed to determine whether the interaction with a specific protein partner might elucidate the biological relevance of DJ-1 missense mutations in cellular physiology. By an yeast two-hybrid (Y2H) screening, we identified TRAF and TNF receptor-associated protein (TTRAP) as a novel DJ-1 interactor.
Human TTRAP was isolated earlier for its ability to bind TNF receptors (TNFR) family members and TNFR-associated factors (TRAFs) inhibiting nuclear factor-κB activation.23 TTRAP was also known as ETS-associated protein II (EAPII).24 The bioinformatic analysis of TTRAP coding sequence revealed an N-terminal ubiquitin-associated (UBA)-like sequence and a C-terminal endonuclease/exonuclease/phosphatase family-like domain.25, 26
Here we show that TTRAP was able to bind M26I and L166P more strongly than WT DJ-1. Furthermore, neuroblastoma cells were protected by TTRAP from apoptosis induced by proteasome impairment. In these conditions, endogenous TTRAP relocalized to a detergent-insoluble fraction and formed juxtanuclear aggresome-like structures. Interestingly, PD-associated missense mutants exerted a pro-apoptotic effect by blocking TTRAP protective activity and inducing c-jun N-terminal-kinase (JNK)- and p38 mitogen-activated protein kinase (MAPK)-mediated cell death.
Results
TTRAP is a novel DJ-1 interactor
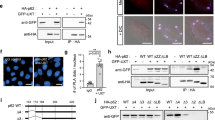
We performed an Y2H screening of a human fetal brain cDNA library to identify novel DJ-1 interaction partners in the brain. After analyzing 107 transformants, we isolated three independent clones encoding for almost the entire open reading frame of T RAF and T NF r eceptor- a ssociated p rotein (NM_016614) (Figure 1a; Supplementary material). We then performed in vitro pull down assays showing that FLAG-TTRAP expressed in mammalian cells interacted with recombinant GST-DJ-1, but not with GST alone (Figure 1b). Using different deletion mutants, we mapped in vitro the interaction to the C-terminal portion of TTRAP (Figure 1b).
TTRAP is a novel DJ-1 interactor. (a) Scheme of TTRAP domains and sequences found in the yeast two-hybrid screening with DJ-1 WT as bait (yeast clones). TTRAP fragments used for pull down assays (ΔN and ΔC) are indicated. (b) TTRAP interacts with DJ-1 in vitro. HEK-293T cells were transfected with FLAG-TTRAP full length, ΔN or ΔC. In vitro binding assay was performed with GST and GST-DJ-1, as indicated. Bound TTRAP was visualized with an anti-FLAG monoclonal antibody. An aliquote of total cell lysate (input) was checked for the expression of transfected protein. Recombinant GST proteins were checked by Ponceau staining. (c) Endogenous DJ-1 interacts with FLAG-TTRAP in human neuroblastoma SH-SY5Y cells. TTRAP was immunoprecipitated from FLAG-TTRAP stably transfected cells with anti-FLAG agarose beads (IP). Control immunoprecipitation (c) was performed on empty vector stable cells. Co-immunoprecipitated endogenous DJ-1 was revealed with anti-DJ-1 antibody. Efficacy of immunoprecipitation was controlled in the same membrane with anti-FLAG antibody. Expression of proteins in cell lysates was verified with anti-FLAG and anti-DJ-1 immunostaining. Molecular weight markers are indicated on the left (kDa). The asterisk (*) indicates an unspecific background band. (d) Immunofluorescence of endogenous TTRAP and endogenous DJ-1 in human neuroblastoma SH-SY5Y cells. Cells were fixed, permeabilized and stained with rabbit polyclonal anti-TTRAP (green) and mouse monoclonal anti-DJ-1 (red) to recognize the endogenous proteins. Nuclei were visualized with DAPI. Colocalization is shown in the merge. (e) TTRAP is expressed in vivo in dopaminergic neurons in Substantia nigra in the mouse brain. Cryo-sections of the ventral midbrain region from C57/B6 mice were taken and endogenous TTRAP was visualized by immunohistochemistry with specific polyclonal antibody. Dopaminergic neurons were identified by staining with anti-TH antibody. Merge shows TTRAP-positive dopaminergic neuron
We then generated human neuroblastoma SH-SY5Y cells stably transfected with FLAG-TTRAP or with empty vector (Supplementary Figure S1; data not shown). After immunoprecipitation with anti-FLAG antibody-coated beads, endogenous DJ-1 was detectable in the TTRAP-bound fraction but not in the negative control (Figure 1c). The binding was weak although specific and reproducible.
Taken together, these results show that TTRAP is a novel DJ-1 interacting protein.
TTRAP is expressed in human neuroblastoma SH-SY5Y cells and in dopaminergic neurons in the Substantia nigra
To study TTRAP expression, we raised a polyclonal anti-TTRAP antibody. Its specificity was confirmed by competition experiments with GST-TTRAP (data not shown) and in SH-SY5Y cells stably transfected with a short hairpin RNA for TTRAP (Supplementary Figure S1).
In neuroblastoma cells, TTRAP was almost exclusively localized to the nucleus where DJ-1 was present as well (Figure 1d). Furthermore, TTRAP was found to be expressed throughout the brain of adult mice (data not shown) with a stronger staining in the dentate gyrus of the hippocampus. In the mesencephalon, it was expressed both in non-dopaminergic and in dopaminergic cells of the Substantia nigra (Figure 1e).
Therefore, TTRAP is expressed in the nucleus of the neurons that are the primary targets of neurodegeneration in PD.
Endogenous TTRAP accumulates in the insoluble fraction of neuroblastoma cells upon oxidative stress and proteasomal impairment
In response to neurotoxic stimuli, PD-associated proteins such as parkin and UCH-L1 undergo a change in subcellular distribution that mimic the formation of aggresome-like structures.6, 7, 27 This is monitored by altered solubility and redistribution into a biochemically defined insoluble fraction.12, 22 Therefore, we tested the distribution of endogenous TTRAP in soluble and insoluble fractions in SH-SY5Y cells treated with PD-mimicking stimuli, such as hydrogen peroxide, 1-methyl-4-phenyl-4-phenylpyridinium ion (MPP+) and the proteasome blocker MG132. Tumor necrosis factor-α (TNF-α) was also monitored for TTRAP involvement in its signal transduction pathway. After treatments, soluble and insoluble fractions were separated and the distribution of endogenous TTRAP was evaluated by western blot analysis. In untreated conditions, TTRAP was almost exclusively present in the soluble fraction (Figure 2). Conversely, after oxidative stress, TTRAP was accumulated in the insoluble fraction. A more potent effect was obtained by MG132. No effect was observed under mitochondrial intoxication or TNF-α treatment.
Endogenous TRAP accumulates in the insoluble fraction in human neuroblastoma SH-SY5Y cells upon proteasomal inhibition. Neuroblastoma SH-SY5Y cells were treated with 1 mM hydrogen peroxide for 1 h, 10 ng/ml TNF-α, 5 mM MPP+ or 5 μM MG132 for 24 h. Control cells were left untreated (UNTR). Cells were lysed, and Triton X-100 soluble and insoluble fractions were separated. The expression of endogenous TTRAP was visualized with anti-TTRAP antibody. Protein loading was controlled by β-actin. Molecular weight markers are indicated for each gel (kDa). Densitometric analysis of protein bands was performed using Adobe Photoshop 7.0 on two independent experiments. Relative expression of endogenous TTRAP was normalized to β-actin (right panels). Data are expressed as the percentage of untreated condition
In summary, TTRAP accumulates in the insoluble fraction upon proteasome impairment.
TTRAP binding to DJ-1 is stronger with M26I and L166P than with WT
To study the binding properties of missense mutations associated with PD, we performed co-immunoprecipitation experiments in HEK-293T (human embryonic kidney) cells (Figure 3a).
TTRAP binding to DJ-1 is potently enhanced with M26I and L166P PD-linked mutants. (a) HEK-293T cells were transfected with FLAG-TTRAP and MYC-DJ-1 WT (W), L166P (L) or M26I (M). Control cells (c) were transfected with MYC-DJ-1 WT and control plasmid. After 24 h transfection, cells were left untreated or incubated with 5 μM MG132 for 16 h, as indicated. Cell lysates were immunoprecipitated (IP) with anti-FLAG agarose beads and bound proteins were revealed by immunoblot (IB) with anti-MYC and anti-FLAG antibodies. An aliquote of each lysate was tested for the expression of TTRAP and DJ-1 proteins. Molecular weight markers are indicated on the left (kDa). HC, heavy chain. (b) Endogenous TTRAP interacts with FLAG-DJ-1 L166P in human neuroblastoma SH-SY5Y cells. L166P (L) or empty vector control (c) stable SH-SY5Y cells were treated with proteasome inhibitor as in panel a, and subjected to IP with anti-FLAG agarose beads. Immunoprecipitates and lysates were analyzed with anti-TTRAP and anti-FLAG antibodies. Molecular weight markers are indicated on the left (kDa). The asterisk (*) indicates an unspecific background band. LC, light chain
When equal amounts of plasmid DNAs were transfected, a lower expression of L166P was repeatedly obtained compared with WT and M26I, as reported in the literature. Therefore, to better measure the binding of TTRAP to WT and DJ-1 mutants, we increased L166P levels by transiently transfecting higher amounts of L166P-encoding plasmid.
Western blot analysis of co-immunoprecipitated proteins showed a weak, although specific, binding of TTRAP to WT DJ-1 in untreated conditions. Conversely, TTRAP displayed a more potent interaction to both PD-linked DJ-1 mutants M26I and L166P. This interaction was not dependent on the concentration of L166P protein because it was confirmed when equal amounts of plasmid DNAs were transfected, leading to lower L166P levels (data not shown).
We then tested whether their interactions might be affected by proteasome impairment. When co-immunoprecipitation experiments were performed in cells treated with MG132, we observed an enhanced binding of TTRAP to WT, M26I and L166P proteins (Figure 3a). These results were further verified by reverse co-immunoprecipitation (data not shown).
Stable transfected SH-SY5Y cells for FLAG-tagged WT-DJ-1 or PD-associated M26I and L166P mutations were then generated. As shown in Supplementary Figure S1, WT and M26I DJ-1s presented similar protein levels, whereas L166P clones showed a much lower level of expression, as reported in the literature for this highly unstable mutant.
After proteasome inhibition, endogenous TTRAP could be consistently co-immunoprecipitated with mutant DJ-1s (Figure 3b).
Taken together, these results prove that TTRAP interacts more strongly with pathogenic forms of DJ-1.
TTRAP protects neuroblastoma cells from MG132-induced apoptosis
As TTRAP solubility is modified by MG132, we studied whether TTRAP overexpression or interference may alter the induction of apoptosis by proteasome impairment.
We performed viability assays on stable SH-SY5Y cells that overexpress TTRAP or an empty vector. After treatment with increasing concentrations, we observed the largest protective effects at 0.25 and 0.5 μM MG132, with a 50% increase in cell viability in TTRAP-overexpressing clones (Figure 4a). These data were confirmed on three independent TTRAP-overexpressing and three vector-containing clones.
TTRAP is protective upon proteasome inhibition. (a) SH-SY5Y cells stably transfected with an empty vector (Control) or with FLAG-TTRAP (TTRAP) were treated for 16 h with the indicated concentration of MG132. Cell viability was measured by MTT assay. Data are expressed as the percentage of untreated cells. Experiments have been repeated on three independent TTRAP and three control clones. *P<0.05. (b) SH-SY5Y TTRAP and control cells were treated as in panel a. Cell lysates were prepared and analyzed by immuno-blot for the presence of p85 PARP fragment. TTRAP overexpression was tested with anti-FLAG antibody. β-Actin staining was used as loading control. The data are representative of three independent experiments. (c) SH-SY5Y control or TTRAP interference (siTTRAP) cells were processed as in panel a and viability was measured by MTT assay. Experiments were repeated on two siTTRAP and two control clones. *P<0.05. (d) siTTRAP and control cells were treated with MG132 and lysates prepared as in panel b. Data are representative of two independent experiments performed on two siTTRAP clones
We also measured apoptosis by following the production of cleaved and enzymatically active p85 fragment of poly-(ADP-ribose) polymerase (PARP). As shown in Figure 4b, TTRAP is essential for reducing PARP activation at lower doses of MG132 treatment. At higher doses, apoptotic pathway is induced independent of TTRAP levels.
We then generated SH-SY5Y stable cell lines with TTRAP-specific siRNA constructs to inhibit the synthesis of endogenous TTRAP protein. Two independent clones were isolated for each of two independent siRNA sequences targeting different regions of TTRAP mRNA (siTTRAP-T1 and siTTRAP-T4) (Supplementary Figure S1).
siTTRAP and control SH-SY5Y cells were treated with increasing concentration of MG132, and cell viability was measured by MTT assay (Figure 4c). As compatible with a protective role of TTRAP in response to proteasome block, T1 and T4 siTTRAP clones displayed a 50% reduction in cellular viability as compared with controls.
siTTRAP cells also showed a strong increase in PARP activity at lower concentrations than controls (0.1 versus 0.25 μM), mirroring the data obtained by overexpressing TTRAP (Figure 4d).
We can then conclude that TTRAP is protective in neuroblastoma cells hampered with proteasome impairment.
Upon MG132 treatment, endogenous TTRAP relocalizes to the cytoplasm forming insoluble aggresomes
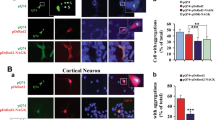
The subcellular localization of endogenous TTRAP was studied after treatment with 0.25 μM MG132 for 16 h, a condition where TTRAP exerts its anti-apoptotic activity.
Endogenous TTRAP partially relocalized to the cytoplasm upon proteasome impairment (Figure 5a), with at least a three-fold increase in cytoplasmic signal (Figure 5b). As MG132 treatment accumulates endogenous TTRAP in the insoluble fraction, we studied the distribution of insoluble TTRAP by immunofluorescence after permeabilization of cells with Triton X-100 before fixation (Figure 5c). Interestingly, endogenous TTRAP from permeabilized cells formed a single large juxtanuclear aggresome-like structure in >90% of cells, whereas no aggresomes were visible in untreated condition.
TTRAP accumulates in the cytoplasm and forms cytoplasmic insoluble aggresomes in response to proteasome impairment. (a) SH-SY5H cells were treated with 0.25 μM MG132 for 16 h or left untreated. Endogenous TTRAP localization was analyzed by confocal immunofluorescence with anti-TTRAP (green). Nuclei were visualized with DAPI. (b) Quantification of TTRAP cytoplasmic staining. A total of 120 cells per condition were counted from three independent experiments. The data indicate mean±S.D. Significance between untreated and treated group was calculated with t-test (P<0.001). (c) SH-SY5H cells were treated as in panel a. Before fixation, cells were permeabilized with Triton X-100 and localization of endogenous TTRAP (green) was analyzed by indirect immunofluorescence. Non-permeabilized cells were used as a reference. Cell shape and morphology was assessed by light transmission, using confocal microscope. Images are representative of three independent experiments. (d) SH-SY5Y cells were treated and permeabilized as in panel c. Aggresomes were visualized by double immunofluorescence with anti-TTRAP (green) and anti-γ-tubulin (red). Colocalization is shown in merge
Immunofluorescence imaging revealed that TTRAP aggregates colocalized with γ-tubulin, a marker of aggresomes. Similar data were obtained in FLAG-TTRAP-overexpressing SH-SY5Y clones stained with an anti-TTRAP or an anti-FLAG antibody (data not shown).
Therefore, proteasome impairment changes the subcellular localization of endogenous soluble TTRAP from the nucleus to the cytoplasm and induces the formation of juxtanuclear insoluble aggresomes containing TTRAP.
TTRAP anti-apoptotic function is inhibited by M26I and L166P mutants, but not by WT DJ-1
To test whether the binding of pathogenic M26I and L166P mutants might have an effect on TTRAP anti-apoptotic functions, we transfected TTRAP- or vector-overexpressing SH-SY5Y cells with equal plasmid concentrations of pathogenic DJ-1 mutants, WT DJ-1 or mock and measured cell death upon proteasomal inhibition. Although in untreated conditions a lower expression of L166P was detectable, the MG132 addition increased mutant DJ-1 levels as reported in the literature. For these experiments, MG132 was used at the concentration (0.25 μM) where TTRAP anti-apoptotic activity was more prominent. Cell death was quantified by three different assays, namely MTT measurements, PARP activation and a single-cell-based assay, to monitor caspase 3 activation by immunofluorescence.
No difference in viability was observed in untreated conditions showing that neither transfection per se nor WT or mutant DJ-1 expression alter cell survival (Figure 6a; UNTR). Upon MG132 treatment, a 40% reduction in viability was observed in all control cells, thus indicating that in these conditions, cells are susceptible to proteasome inhibition as expected and no further toxicity was given by mutant DJ-1 proteins (Figure 6a; MG132). In TTRAP-overexpressing cells, we observed, as shown earlier, an increase in cell viability. No effects were detected when WT DJ-1 was co-expressed. Conversely, M26I and L166P mutants significantly inhibited (P<0.002) TTRAP protective activity (Figure 6a). Analogous results were obtained on additional TTRAP-overexpressing and control clones as well as using FLAG-tagged DJ-1 constructs (data not shown). As shown in Figure 6b, data obtained measuring PARP activation were comparable with those obtained in MTT assays. Importantly, p85 PARP accumulation was repeatedly observed when L166P and M26I protein levels were lower than those of WT DJ-1 (data not shown), a condition believed to be occurring in vivo.
TTRAP protective activity is blocked by PD-linked DJ-1 M26I and L166P mutants. (a) SH-SY5Y cells stably transfected with an empty vector (Control) or with FLAG-TTRAP (TTRAP) were transfected with control plasmid (c) or MYC-DJ-1 WT (W), L166P (L) and M26I (M) as indicated. After transfection, cells were treated with 0.25 μM MG132 for 16 h and used for MTT assays. Data are expressed as the percentage of control untreated cells. Standard deviations are indicated and calculated on triplicates. Data have been confirmed on other two independent TTRAP and control clones. *P<0.002; NS, nonsignificant. (b) SH-SY5Y TTRAP and control cells were transfected and treated as in panel a. Cell lysates were prepared and analyzed for the expression of p85 PARP fragment, FLAG-TTRAP and MYC-DJ-1, as indicated. Equal loading was verified with β-actin staining. Molecular weights are indicated on the left (kDa). Data have been confirmed on two independent TTRAP and control clones. (c) SH-SY5Y TTRAP and control cells were transfected as indicated and treated with MG132. Double immunofluorescence was performed with anti-MYC and anti-active caspase-3 antibodies. Data were collected from 100 cells per condition from two independent experiments. *P<0.002
We then set up a single cell-based assay to measure caspase 3 activation. SH-SY5Y control or FLAG-TTRAP-overexpressing cells were transiently transfected with WT, M26I or L166P DJ-1, and the induction of apoptosis upon MG132 treatment was followed by immunofluorescence with an antibody specific for active caspase 3. Mock-transfected cells were used as an additional control. As expected, no signal for active caspase 3 was measurable in untreated cells. Upon proteasome inhibition, a clear signal corresponding to active apoptosis was measured (see Materials and Methods). Figure 6c shows that TTRAP was protective upon proteasome inhibition and overexpression of WT DJ-1 did not change TTRAP protective activity. In contrast, M26I and L166P abolished TTRAP-mediated anti-apoptotic effect.
Mutant DJ-1 proteins exert their pro-apoptotic function through the activation of JNK and p38-MAPK pathways
As MAPKs are known mediators of apoptosis triggered by proteasome impairment,28 we monitored whether the anti-apoptotic property of TTRAP was able to inhibit JNK activation.
Figure 7a shows that JNK phosphorylation was not detectable in TTRAP-overexpressing clones, whereas it was visible in control cells after MG132 treatment. Importantly, when M26I and L166P were expressed in TTRAP-overexpressing cells, they were both able to rescue MG132-triggered JNK activation (Figure 7b). To our knowledge, this is the first time that two PD-associated missense mutations share a common effect on a signal transduction pathway. Furthermore, these data prove that this effect depends on the expression of TTRAP.
M26I and L166P blockade of TTRAP protective activity is mediated by the pro-apoptotic JNK and p38 pathways. (a) SH-SY5Y cells stably transfected with an empty vector (c) or with FLAG-TTRAP (TT) were left untreated (UNTR) or treated for 16 h with 0.25 μM MG132, as indicated. Cell lysates were prepared and analyzed for the expression of phosphorylated JNK1/2 (pJNK1/2). The expression of TTRAP in stable cells was verified with anti-FLAG. Equal loading was confirmed with β-actin staining. Molecular weights are shown (kDa). (b) SH-SY5Y cells stably transfected with an empty vector (Control) or with FLAG-TTRAP (TTRAP) were transfected with control plasmid (c) or MYC-DJ-1 WT (W), L166P (L) and M26I (M) as indicated. After transfection, cells were left untreated (UNTR) or treated for 16 h with 0.25 μM MG132. Cell lysates were prepared and analyzed for the expression of pJNK1/2, FLAG-TTRAP, MYC-DJ-1 and β-actin. (c) TTRAP and control cells were transfected as in panel b. After transfection, cells were left untreated (UNTR) or treated for 16 h with 0.25 μM MG132 alone (MG132) or in combination with 10 μM JNK1/2 inhibitor SP600125 together with 20 μM p38-inhibitor SB203580 (+SP/+SB). Cell lysates were prepared and analyzed as in panel b. (d) Cells were transfected and treated with MG132 with each inhibitor alone, as indicated. All data are representative of three independent experiments performed on two independent TTRAP and control clones
This led us to monitor M26I and L166P-dependent apoptosis in the presence of specific inhibitors of JNK (SP600125) and p38 kinases (SB203580). The co-administration of SP600125 and of SB203580 was not able to block MG132-mediated apoptosis in control cells (Figure 7c). However, TTRAP-dependent M26I and L166P pro-apoptotic activities were completely abolished after SP600125 and SB203580 co-administration proving that PD-associated missense mutations specifically trigger a combined JNK- and p38-mediated apoptotic pathway in a TTRAP-dependent manner. No effects were observed when a single drug was added (Figure 7d).
These data prove that the ‘gain-of-function’ effect exerted by M26I and L166P occurs by inducing JNK- and p38 MAPK-mediated apoptosis.
Discussion
The description of DJ-1 interactors is important for identifying biological functions that may be involved in DJ-1 neuroprotective activity and be impaired in PD. In vivo and in vitro models had shown that lack of DJ-1 impairs cellular responses to neurotoxic stimuli. Mutants such as L166P were unable to rescue cellular homeostasis and this behavior was often linked to the loss of interaction with DJ-1 physiological partners. In this context, DJ-1 binding was important for its chaperone activity on α-synuclein and for sequestering Daxx into the nucleus, preventing Daxx-mediated ASK1 activation.29 Furthermore, WT DJ-1 was able to de-repress promoters by binding to PSF and block apoptosis, whereas L166P, M26I and D149A mutants were not.11
Therefore, the ‘state-of-the-art’ view says that DJ-1 deletions and missense mutations all share a common phenotype based on a ‘loss of function.’ The failure of all missense mutations to protect neurons from exogenous insults may reside in a decreased ability to form dimers, as required for an active DJ-1, and to interact with protein partners.
The ‘loss-of-function’ hypothesis stems from the biochemical and structural properties of L166. This mutant presents a structural rearrangement that interferes with dimer formation favoring protein instability.20 However, mutants such as M26I are more stable than L166P, showing protein levels comparable or slightly lower than WT. Furthermore, recent computational and biochemical studies have shown that M26I dimerizes.18
Therefore, a common pathological mechanism shared by all mutants is still lacking. Most importantly, as most of the experiments have been performed on the highly unstable L166P protein, more studies are needed on dimer-forming, more stable DJ-1 mutants such as M26I.
Interestingly, missense mutations may also present ‘gain-of-function’ properties. Gene profiling experiments in DJ-1 KO and in L166P-NIH3T3 cells30 have shown that L166P affects the expression of a different repertory of genes. Interestingly, L166P upregulated the tau promoter, whereas WT DJ-1 was acting as a repressor. An L166P-mediated dominant-negative effect on the antioxidant WT DJ-1 activity has also been postulated.16 Unfortunately, these works did not present any data on dimer-forming, more stable DJ-1 mutants such as M26I.
Any ‘gain-of-function’ property probably involves some new protein–protein interactions. To our knowledge, the only examples of specific binding shared by M26I and L166P DJ-1 mutants occurred with parkin.22
Here we show that M26I and L166P share an increased interaction with TTRAP. This binding occurs in the absence of neurotoxic stimuli proving the existence of an altered DJ-1 protein networks in the presence of PD-associated missense mutations. As for parkin, this interaction is strongly enhanced after proteasome impairment. As M26I and L166P higher protein levels cannot fully account for TTRAP stronger binding, post-translational modifications of TTRAP or for mutant DJ-1s might be induced by proteasome block modifying the strength of the interaction. Further studies are needed to address this interesting issue.
Most importantly, M26I and L166P block the protective activity of TTRAP to MG132 treatment and induce a TTRAP-dependent pro-apoptotic pathway involving JNK and p38 MAPKs, a signaling system that plays a central role in dopaminergic cell death and PD pathogenesis.31
M26I and L166P were not toxic per se. They exerted their pro-apoptotic function only in the presence of TTRAP. Importantly, their activity was not simply due to a sequestration of an anti-apoptotic protein, as M26I- and L166P-expressing cells did not die in the presence of kinase inhibitors in a similar manner to controls. Therefore, M26I and L166P have conferred TTRAP a novel signaling property that allows an otherwise anti-apoptotic protein to induce cell death through the activation of stress kinases. M26I and L166P thus exert their potential ‘gain of function’ by their binding to TTRAP.
Many mechanisms may be proposed on how M26I and L166P may induce a gain of pro-apoptotic function in TTRAP and how TTRAP may partially block MG132-induced cell death. TTRAP-mediated cell signaling may involve its enzymatic activity as well as its protein interaction network. TTRAP belongs to a large family of endonuclease/exonuclease/phosphatase proteins with a plethora of diverse biochemical activities.26 Its significant homology with Ape1 and interaction with ets family members may suggest a potential role in transcription.24
Upon proteasome impairment, TTRAP relocalizes to the cytoplasm and forms aggresome-like structures. Soluble cytoplasmic TTRAP may indeed be involved in cell signaling events. TTRAP has been shown earlier to bind the intracellular domain of TNF receptor family members and their associated proteins TRAFs, with TRAF6 being the strongest partner.23 Furthermore, TTRAP acted as an inhibitor of nuclear factor-κB activation. This role seems very unlikely in these experimental settings for the well-known suppression of the nuclear factor-κB pathway by MG132. Therefore, the molecular events associated with the regulation of the MAPK pathways by TTRAP and missense mutants remain unclear. A deeper understanding of TTRAP protein network will be needed.
A portion of TTRAP accumulates in insoluble, cytoplasmic, aggresome-like structures. Although the formation of aggresomes by endogenous proteins is a rare phenomenon, proteins involved in neurodegenerative diseases are often able to do so under stress conditions or when mutated. In this context, parkin and UCH-L1 are among the most well-known aggresome-prone molecules.6, 7, 32 It is therefore interesting that endogenous TTRAP may form aggresome-like structures upon proteasome inhibition. These structures have been recently linked to non-degradative Lys-63 ubiquitination and to the action of HDAC6.33, 34 More recently, L166P mutants but not WT DJ-1 have been shown to accumulate in insoluble aggresome-like structures upon MG132 after parkin and HDAC6 binding.35 It will be interesting to investigate whether TTRAP plays a role in HDAC6-dependent aggresome formation and whether HDAC6 activity may be extended beyond L166P mutant.
Our data suggest that DJ-1 missense mutations may combine two pathological insults: a ‘loss of function’ with a decreased ability to cope with oxidative stress leading to mitochondrial dysfunction and a ‘gain of function’ with a TTRAP-dependent induction of MAPKs-mediated apoptosis. Its relevance in PD will depend on the level of these proteins in vivo.
We are aware of the limitations in the use of human neuroblastoma cell lines to recapitulate dysfunctions that occur in dopaminergic neurons of the human brain. Nonetheless, it must be noted that to the best of our knowledge, no investigation of DJ-1 missense mutations has been carried out in post-mortem brains of PD patients and in knock-in animal models. Furthermore, experiments in the human neuroblastoma SH-SY5Y cell line have been central in unveiling DJ-1 protective activity against neuronal apoptosis.
Therefore, the combined ‘loss- and gain-of-functions’ hypothesis would represent a provocative example for mechanisms of neurodegeneration, as already proven for Huntington's disease36 and amyotrophic lateral sclerosis.37
Materials and Methods
Constructs
Full-length human TTRAP sequence was obtained by RZPD (Germany). Open reading frame was subcloned by PCR into pCDNA3-2XFLAG (EcoRI/XbaI) and pCS2-6XMYC (EcoRI/XbaI) plasmids. Expression vectors encoding for WT, M26I and L166P DJ-1 were described earlier.18 Two different oligonucleotide sequences were selected for the knockdown TTRAP expression using siRNA Target Finder software (Invitrogen). The hairpin-encoding oligonucleotides were cloned into the pSuperior vector (Invitrogen). Details are available on Supplementary material.
Cell culture and transfections
HEK-293T (Sigma) and human neuroblastoma SH-SY5Y cells (ATCC) were maintained in culture as suggested by vendors (see Supplementary materials). Transfections of HEK cells were performed with the standard calcium phosphate method (transfection efficiency rate of about 60–70%). As SH-SY5Y cells could be poorly transfected with the calcium phosphate method (5–10% efficiency), cells were transfected with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions, to achieve 60–70% transfection efficiency.
We generated SH-SY5Y stable cell lines by transfecting linearized constructs with Lipofectamine 2000 and selecting for positive clones in the presence of 300 μg/ml of Geneticin (Invitrogen). Control cells were generated by transfecting an empty vector and following the same selection procedure. Individual clones were confirmed by western blot analysis, immunofluorescence and quantitative real-time PCR. Constructs encoding for FLAG-TTRAP, FLAG-DJ-1 WT, M26I and L166P and pcDNA3-FLAG empty vectors were used. Stable neuroblastoma cells were also generated with knocked down TTRAP expression upon transfection with pSuperior-T1 and -T4.
Pull down, immunoprecipitation and western blot analysis
For in vitro binding, HEK-293T cells were transfected with the indicated vectors and further processed after 36 h. Cells were lysed with pull-down lysis buffer (Supplementary material), and equal amount of cell lysate was incubated with GST or GST-DJ-1 proteins bound to glutathione-sepharose beads (GE Healthcare) After extensive washing, bound proteins were eluted in 2 × SDS sample buffer, boiled and analyzed by western blot.
For co-immunoprecipitation experiments, cells were lysed in IP or endo-IP buffers (Supplementary material). Cellular lysates were incubated with anti-FLAG agarose beads (Sigma) or with the appropriate antibody. After washing, immunoprecipitated proteins were eluted with 2 × SDS sample buffer, boiled and analyzed by western blot. The following antibodies were used: anti-FLAG 1 : 2000 (Sigma), anti-MYC 1 : 4000 (Cell Signaling), anti-TTRAP 1 : 1000 (purified from immunized rabbits), anti-DJ-1 1 : 1000 (purified from immunized rabbits), anti-p85-PARP 1 : 500 (Cell Signaling) and anti-β-actin 1 : 5000 (Sigma).
Immunocytochemistry and immunohistochemistry
For immunofluorescence experiments, SH-SY5Y cells were fixed in 4% paraformaldehyde, and indirect immunofluorescence was performed following standard protocols (Supplementary material). We used anti-FLAG (1 : 1000), anti-MYC (1 : 2000), anti-TTRAP (1 : 100), anti-DJ-1 1 : 100 (StressGene) and anti-cleaved Caspase 3 1 : 100 (Cell Signaling) antibodies. To detect, Alexa Fluor-488 or Alexa Fluor-594 (Invitrogen)-labeled anti-mouse or anti-rabbit antibodies were used. Nuclei were visualized with DAPI (1 μg/ml).
To detect insoluble TTRAP, immunocytochemistry was performed as described earlier.38
For immunohistochemistry, 8- to 12-weeks old C57/B6 mice (Jackson Laboratories) were fully anesthetized and perfused with PBS followed by 4% paraformaldehyde in PBS. Brains were dissected, post-fixed in 4% paraformaldehyde and cryo-protected in 30% sucrose overnight. Coronal cryo-sections were prepared, blocked in IHC-blocking solution (Supplementary material) and stained with anti-TTRAP and anti-tyrosine hydroxylase (Chemicon). Nuclei were visualized with DAPI.
All images were collected using a confocal microscope (LEICA TCS SP2). For quantification of cytoplasmic TTRAP, high-resolution images were analyzed with ImageJ software. A randomly selected area was traced in the cytoplasm, and TTRAP was fluorescence measured. Background fluorescence was quantified from an area placed outside the cells and was subtracted for each signal.
RNA isolation, reverse transcription and qPCR
Total RNA was isolated using the TRIZOL reagent (Invitrogen) following the manufacturer's instructions. Single-strand cDNA was obtained from 1 μg of purified RNA using the iSCRIPT™ cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's instructions. Quantitative RT-PCR (qPCR) was performed using SYBR-Green PCR Master Mix (Applied Biosystems) and an iCycler IQ Real-Time PCR System (Bio-Rad). Expressions of DJ-1 and TTRAP were analyzed using specific oligonucleotides (Supplementary material).
Cell viability and apoptosis
Cell viability was measured using MTT (Sigma) assays following the manufacturer's instructions and reading absorbance at 570 nm using a standard spectrophotometer.
Apoptosis was assessed as mean of caspase-mediated cleavage of PARP by western blot analysis of cell lysates with PARP p85 fragment-specific antibody (Cell Signaling). Single-cell apoptosis was followed by immunofluorescence with cleaved caspase 3-specific antibody (1 : 100; Cell Signaling).
Statistical analysis
All experiments were repeated in duplicate or more. For stably transfected cells, at least two independent clones were used for each cell line in all experiments. Data represent the mean ±S.D.. When necessary, each group was compared individually with reference control group using Student's t-test (Microsoft Excel software).
Abbreviations
- PD:
-
Parkinson's disease
- TTRAP:
-
TRAF and TNF receptor-associated protein
- UCH-L1:
-
ubiquitin C-terminal hydrolase-L1
References
Thomas B, Beal MF . Parkinson's disease. Hum Mol Genet 2007; 16 (Spec No. 2): R183–R194.
Betarbet R, Sherer TB, Greenamyre JT . Ubiquitin–proteasome system and Parkinson's diseases. Exp Neurol 2005; 191 (Suppl 1): S17–S27.
McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW . Altered proteasomal function in sporadic Parkinson's disease. Exp Neurol 2003; 179: 38–46.
Petrucelli L, O'Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron 2002; 36: 1007–1019.
Rideout HJ, Lang-Rollin IC, Savalle M, Stefanis L . Dopaminergic neurons in rat ventral midbrain cultures undergo selective apoptosis and form inclusions, but do not up-regulate iHSP70, following proteasomal inhibition. J Neurochem 2005; 93: 1304–1313.
Muqit MM, Davidson SM, Payne Smith MD, MacCormac LP, Kahns S, Jensen PH et al. Parkin is recruited into aggresomes in a stress-specific manner: over-expression of parkin reduces aggresome formation but can be dissociated from parkin's effect on neuronal survival. Hum Mol Genet 2004; 13: 117–135.
Ardley HC, Scott GB, Rose SA, Tan NG, Robinson PA . UCH-L1 aggresome formation in response to proteasome impairment indicates a role in inclusion formation in Parkinson's disease. J Neurochem 2004; 90: 379–391.
Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003; 299: 256–259.
Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci USA 2007; 104: 14807–14812.
Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA . The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc Natl Acad Sci USA 2003; 100: 9256–9261.
Xu J, Zhong N, Wang H, Elias JE, Kim CY, Woldman I et al. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum Mol Genet 2005; 14: 1231–1241.
Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A . DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol 2004; 2: e362.
Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H . Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun 2003; 312: 1342–1348.
Bonifati V, Oostra BA, Heutink P . Linking DJ-1 to neurodegeneration offers novel insights for understanding the pathogenesis of Parkinson's disease. J Mol Med 2004; 82: 163–174.
Goldberg MS, Pisani A, Haburcak M, Vortherms TA, Kitada T, Costa C et al. Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial Parkinsonism-linked gene DJ-1. Neuron 2005; 45: 489–496.
Kim RH, Smith PD, Aleyasin H, Hayley S, Mount MP, Pownall S et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci USA 2005; 102: 5215–5220.
Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, Wes PD et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson's disease. Curr Biol 2005; 15: 1572–1577.
Herrera FE, Zucchelli S, Jezierska A, Lavina ZS, Gustincich S, Carloni P . On the oligomeric state of DJ-1 protein and its mutants associated with Parkinson Disease. A combined computational and in vitro study. J Biol Chem 2007; 282: 24905–24914.
Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA . Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry 2008; 47: 1381–1392.
Macedo MG, Anar B, Bronner IF, Cannella M, Squitieri F, Bonifati V et al. The DJ-1L166P mutant protein associated with early onset Parkinson's disease is unstable and forms higher-order protein complexes. Hum Mol Genet 2003; 12: 2807–2816.
Baulac S, LaVoie MJ, Strahle J, Schlossmacher MG, Xia W . Dimerization of Parkinson's disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol Cell Neurosci 2004; 27: 236–246.
Moore DJ, Zhang L, Troncoso J, Lee MK, Hattori N, Mizuno Y et al. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum Mol Genet 2005; 14: 71–84.
Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N et al. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J Biol Chem 2000; 275: 18586–18593.
Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R . EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene 2003; 22: 2699–2709.
Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K et al. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 2005; 435: 1257–1261.
Rodrigues-Lima F, Josephs M, Katan M, Cassinat B . Sequence analysis identifies TTRAP, a protein that associates with CD40 and TNF receptor-associated factors, as a member of a superfamily of divalent cation-dependent phosphodiesterases. Biochem Biophys Res Commun 2001; 285: 1274–1279.
Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM . Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J Biol Chem 2004; 279: 4625–4631.
Meriin AB, Gabai VL, Yaglom J, Shifrin VI, Sherman MY . Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem 1998; 273: 6373–6379.
Junn E, Taniguchi H, Jeong BS, Zhao X, Ichijo H, Mouradian MM . Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci USA 2005; 102: 9691–9696.
Nishinaga H, Takahashi-Niki K, Taira T, Andreadis A, Iguchi-Ariga SM, Ariga H . Expression profiles of genes in DJ-1-knockdown and L 166 P DJ-1 mutant cells. Neurosci Lett 2005; 390: 54–59.
Ferrer I, Blanco R, Carmona M, Puig B, Barrachina M, Gomez C et al. Active, phosphorylation-dependent mitogen-activated protein kinase (MAPK/ERK), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), and p38 kinase expression in Parkinson's disease and dementia with Lewy bodies. J Neural Transm 2001; 108: 1383–1396.
Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay SP et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin's protective function. Hum Mol Genet 2005; 14: 3885–3897.
Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP . The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003; 115: 727–738.
Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 2008; 17: 431–439.
Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 2007; 178: 1025–1038.
Cattaneo E, Rigamonti D, Goffredo D, Zuccato C, Squitieri F, Sipione S . Loss of normal huntingtin function: new developments in Huntington's disease research. Trends Neurosci 2001; 24: 182–188.
Sau D, De Biasi S, Vitellaro-Zuccarello L, Riso P, Guarnieri S, Porrini M et al. Mutation of SOD1 in ALS: a gain of a loss of function. Hum Mol Genet 2007; 16: 1604–1618.
Rubbi CP, Milner J . Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 2000; 19: 85–96.
Acknowledgements
We are indebted to Dr. Patrizia Rizzu (University Medical Center, Amsterdam, The Netherlands) for kindly providing human DJ-1 cDNA clone; to Dr. Francesca Persichetti (SISSA, Trieste, Italy) for LexA-Rrs1 plasmid and for carefully reading the manuscript and to Dr. Licio Collavin (University of Trieste, Italy) for antibodies, constructs and helpful discussions. We thank all the members of the SG lab for thought-provoking discussions and help. This work was supported by the Telethon Grant GGP06268, by The Giovanni Armenise-Harvard Foundation and by the Italian Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by N Bazan
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Zucchelli, S., Vilotti, S., Calligaris, R. et al. Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson's disease-associated DJ-1 missense mutations. Cell Death Differ 16, 428–438 (2009). https://doi.org/10.1038/cdd.2008.169
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.169
Keywords
This article is cited by
-
The E3 Ubiquitin Ligase TRAF6 Interacts with the Cellular Prion Protein and Modulates Its Solubility and Recruitment to Cytoplasmic p62/SQSTM1-Positive Aggresome-Like Structures
Molecular Neurobiology (2022)
-
DJ-1 modulates the unfolded protein response and cell death via upregulation of ATF4 following ER stress
Cell Death & Disease (2019)
-
Upregulated Parkin expression protects mitochondrial homeostasis in DJ-1 konckdown cells and cells overexpressing the DJ-1 L166P mutation
Molecular and Cellular Biochemistry (2014)
-
SUMO and Parkinson’s Disease
NeuroMolecular Medicine (2013)
-
L166P mutant DJ-1 promotes cell death by dissociating Bax from mitochondrial Bcl-XL
Molecular Neurodegeneration (2012)