Abstract
Background:
Aldehyde dehydrogenase (ALDH) has been widely used as a marker of cancer stem cells (CSCs). However, the ALDH family includes 19 members, and the most relevant isoforms and their biological functions in cancer biology are still controversial.
Methods:
We examined ALDH enzyme activity and the mRNA expression of 19 ALDH members in 58 human cell lines. The biological effect and mechanism of knocking down ALDH1A3 with siRNA and shRNA in cell lines were explored. Finally, the relationship between ALDH1A3 and CXCR4 was analysed in a large panel of cell lines.
Results:
ALDH1A3 is the key isoform that contributed to Aldefluor positivity in cell lines. Knocking down ALDH1A3 in different cancer cells conferred opposite phenotypes due to differential effects on CXCR4 expression. There was a significant negative correlation between ALDH1A3 and CXCR4 in 58 human cell lines.
Conclusions:
ALDH1A3 was the main contributor to Aldefluor positivity in human cell lines, and its contrasting effects might arise from differences in CXCR4 expression.
Similar content being viewed by others
Main
Aldehyde dehydrogenases (ALDHs) are members of a family of NAD-dependent enzymes that catalyse the oxidation of aldehydes to acids. ALDHs are localised in the cytoplasm, mitochondria or nucleus and have been implicated in a wide variety of biological processes, including the detoxification of exogenous aldehydes and the metabolism of vitamin A, alcohol and ROS (Marchitti et al, 2008). In addition, ALDH activity has been widely used as a marker of cancer stem cells (CSCs). However, the ALDH enzyme family includes 19 members, and the currently available commercial assays identify ALDH+ cells as those actively metabolising BODIPY-aminoacetaldehyde (Aldefluor reagent), which does not distinguish the specific ALDH isoforms (Morgan et al, 2015). Different ALDH isozymes might contribute to the elevated ALDH activity in different types of cancer. ALDH1A1 was thought to be the key isozyme responsible for Aldefluor activity (Xu et al, 2015). However, recently, other ALDH isoforms, such as ALDH1A3 (Chen et al, 2016), ALDH1B1 (Singh et al, 2015), ALDH3A1 (Yan et al, 2014) and ALDH7A1 (Marcato et al, 2011a) have been reported to be the cause of ALDH Aldefluor activity in various tissues and cancers. In particular, ALDH1A3 is receiving more attention because of its critical role in breast (Marcato et al, 2015), lung (Shao et al, 2014) and brain cancers (Cheng et al, 2016).
There are many papers about the abnormal expression of ALDH1A3 closely associated with the biological properties in various tumours, but the data regarding the effect of ALDH1A3 on modulating the malignant phenotype are contradictory, even in studies that focused on clinical tumour specimens of the same origins. For example, in head and neck squamous cell carcinoma, normal tissue and cancer adjacent tissue showed significantly higher ALDH1A3 intensity compared to the tumour tissues (Kurth et al, 2015). In addition, in 455 non-small cell lung cancer specimens, high expression of ALDH1A3 was associated with well-differentiated tumours and better overall survival (Shao et al, 2014). However, ALDH1A3 expression was increased in clinical high-grade glioma compared with low-grade glioma or normal brain tissue (Mao et al, 2013), and ALDH1A3 was modulated by stat3 (Shao et al, 2014; Canino et al, 2015). Depletion of ALDH1A3 promoted the accumulation of apopto-genic aldehydes leading to apoptosis and tumour growth inhibition (Perez-Alea et al, 2017), while Marcato reported an opposing tumour growth effect of ALDH1A3 in breast cancer cells, that was speculated to result from differential retinoic acid (RA) signalling (Marcato et al, 2015).
Here, we report that ALDH1A3 is the key isozyme that is functionally important for maintaining ALDH activity in a panel of 58 human cell lines. Furthermore, the potential function and underlying mechanisms that ALDH1A3 exerts on the colon cancer malignancy were investigated.
Materials and methods
Cell lines
All cell lines used in this study were obtained from the Cell Resource Center, Peking Union Medical College (which is the headquarters of the National Infrastructure of Cell Line Resource, NSTI) and cultured according to the recommended methods. The cell lines were checked and found to be free of mycoplasma contamination using PCR and culture. The identity of the cell lines was authenticated with short tandem repeat profiling (FBI, CODIS). All the results can be viewed on the website (http://cellresource.cn).
Aldefluor assay and separation of ALDH+ cell populations
The Aldefluor kit (Stem Cell Technologies, Vancouver, BC, Canada) was used to analyse and identify cell populations with high ALDH enzymatic activity. Briefly, 2 × 106 cells ml−1 were suspended in Aldefluor assay buffer containing the ALDH substrate BODIPY-aminoacetaldehyde and incubated at 37 °C for 45 min. For each sample, cell aliquots were incubated with or without 50 mM diethylaminobenzaldehyde (DEAB), an ALDH-specific inhibitor. Then, the cells were analysed on an Accuri C6 Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). The difference of the mean fluorescence intensity between the test cells (without DEAB) and control cells (with DEAB) was quantified for accurate Aldefluor activity in the 58 cell lines. For fluorescence-activated cell sorting (FACS), a FACSDiVa flow cytometer (Becton Dickinson) was used to sort subpopulations of colorectal cancer cells based on ALDH activity.
RNA isolation, reverse transcription, real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was performed with a PrimeScript RT Master Mix kit (Takara, Dalian, China). Real-time PCR was performed in a StepOnePlus system using the following program: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Each reaction was carried out in triplicate. To calculate the relative expression levels, we used the 2-ΔΔCT-method. All primer sequences are presented in Supplementary Table 1. GAPDH mRNA was used as an endogenous control.
Western blotting
Protein samples were separated by 10% SDS–PAGE (Beyotime, Shanghai, China) and transferred onto NC membranes (Pall, Stevenage, UK). For immuno-detection, the following primary antibodies were used: anti-ALDH1A3 (ab129815, 1 : 1000, Abcam, Cambridge, UK), anti-ALDH1A1 (GTX100043, 1 : 2000, GeneTex, Irvine, CA, USA), anti-CXCR4 (GTX22074, 1 : 1000, GeneTex) and anti-GAPDH (1 : 2000, Santa Cruz, Dallas, TX, USA). After incubation overnight at 4 °C, blots were washed and incubated with secondary antibodies for 1 h at room temperature. ECL Super Signal Substrate (Pierce, Appleton, WI, USA) was applied for visualisation of the bands, and the optical densities of the bands in each blot were analysed using Quantity One 1-D Analysis System (Bio-Rad, Hercules, CA, USA).
Cell growth and cell cycle analysis
Cell growth was analysed using methyl thiazole tetrazolium (MTT, Sigma, St Louis, MO, USA) or Cell Counting Kit-8 (CCK8) (Bestbio, China) assays. methyl thiazole tetrazolium assays were performed as previously described (Zhang et al, 2013), and the quantification of viable cells was measured as the optical density (OD) at 570 nm. For CCK8 assays, 10 μl of CCK8 solution was added to each well and incubated for an additional 3 h at 37 °C, and then, the OD of the supernatants was measured at 450 nm.
For cell cycle analysis, fixed cells were resuspended in PBS containing 25 μg ml−1 RNase and incubated for 1 h at 37 °C, and then, the cells were stained with 50 μg ml−1 PI for 15 min at 4 °C. The data were analysed using ModFit LT 3.2.
In vitro invasion assays
In vitro invasive ability was assayed with Transwell inserts containing 8.0-μm pores (Costar, Cambridge, MA, USA) coated on the bottom with Matrigel (354277, BD). Cells were placed in the upper chamber in 0.2 ml of serum-free medium, and the lower chamber was filled with 0.5 ml of conditioned medium (serum-free supernatant of NIH/3T3 cells with 20% FBS). Cells were incubated for 48 h, and the invasive cells on the lower surface of the membrane were fixed in 4% paraformaldehyde and stained with crystal violet for 30 min. The cells in nine randomly selected microscopic fields per well (× 20) were counted.
siRNA silencing and shRNA stable expression
Small interfering RNA (siRNA) was used to knockdown ALDH1A1 and ALDH1A3 expression in cancer cells. With the exception of si-s30, which targeted ALDH1A3 and was purchased from Invitrogen (4390824), all siRNAs were obtained from Ribo Bio Co. All the siRNA sequences are listed in Supplementary Table 1, and Ncontrol_05815 (Ribo Bio, Guangzhou, China) was used as a negative universal control (siNC). Transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Two shRNA retroviruses targeting human ALDH1A3 (sh-A33/sh-s30) and a control shRNA (sh-scr) retrovirus were constructed using a 3-plasmid packaging system. Briefly, the pSIREN-RetroQ Vector expressing the shRNA sequence was cotransfected into 293T cells together with vectors expressing the gag and vsvg genes using FugeneHD (Roche, Mannheim, Germany). After 48 h of incubation, the viral supernatant was collected and used to transduce cells in the presence of 10 μg ml−1 polybrene (Sigma Aldrich, St Louis, MO, USA). The infected cells were selected with puromycin (1 μg ml−1) for 1 week to generate stable shRNA-expressing cancer cells and then continuously cultured with 0.5 μg ml−1 puromycin.
Transient CXCR4 transfection
The CXCR4 gene fragments were amplified from human genomic DNA and cloned into pcDNA3.1 plasmid, and the plasmid was transfected into cells for transient expression of CXCR4.
Statistical analysis
All statistical calculations were performed using GraphPad Prism Software Version 5.0. Correlation analyses of ALDH Aldefluor activity and the mRNA expression of 19 isozymes and the correlations between ALDH1A1, ALDH1A3, CXCR4 and CXCR7 were determined by Spearman rank correlation tests. The data are presented as the mean±s.d. All P-values were two-tailed, and P<0.05 was considered statistically significant.
Results
ALDH1A3 is the key ALDH isozyme responsible for Aldefluor activity in human cell lines
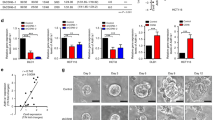
We examined ALDH enzyme activity using an Aldefluor assay in 58 popular human cell lines. The positive cells in large exhibited one population rather than two distinct positive/negative bulks, and the ALDH+ ratio differed from 0.0 to 85.7% in flow cytometry (Figure 1A and Supplementary Table 2). Multiple ALDH isoforms could contribute to the Aldefluor fluorescence. Therefore, we checked the mRNA expression of 19 ALDH family members in the panel of cell lines using real-time PCR, and the results suggested different expression patterns of these ALDHs (Supplementary Figure 1). For example, the mRNA levels of 3A2, 7A1, 9A1 and 18A1 were readily detected at a relatively stable high level in all the cells, while 1L1, 1L2, 8A1 and 16A1 exhibited a much lower expression. On the other hand, expression of 1A1, 1A2, 1A3 and 3A1 was highly variable in different cell lines. Among the 19 isoforms studied, a positive correlation was only observed between ALDH1A3 mRNA expression and the Aldefluor activity (r=0.53, P<0.01), suggesting that ALDH1A3 is the key ALDH isozyme that is functionally important for maintaining ALDH+ cells (Figure 1B and C). The 58 human cell lines were further divided into eight groups according to their tissue origin, and the relationship between ALDH1A3 expression and the Aldefluor activity was especially close in pulmonary cancers, colorectal cancers, breast and female reproductive system cancers (Figure 1D). We also confirmed ALDH1A3 and ALDH1A1 protein expression in eight colon cancer cell lines, two pulmonary cancer cell lines and four gastric carcinoma cell lines (Figure 1E), which correlated with their mRNA level (Supplementary Figure 2).
ALDH1A3 is the key isotype responsible for ALDH+ cells in 58 human cell lines. (A) Representative photomicrographs of Aldefluor assays in five human cancer cell lines. (B) and (C) Spearman analyses of the mRNA expression of 19 ALDH isoforms by qPCR and the Aldefluor activity obtained from Aldefluor assays in 58 human cell lines. (D) Correlation between mRNA expression of ALDH1A3 and Aldefluor activity in cells from different tissues of origin. (E) Western blotting analysis of ALDH1A1 and ALDH1A3 in eight colorectal cancer cell lines, two lung cancer cell lines and four gastric cancer cell lines.
The ALDHhigh/+ subpopulation of colon cancer cells has CSC properties
ALDH can be used as a CSC marker in many tumours. Thus, we verified whether ALDH could enrich colon CSCs in colon cancer cell lines in vitro. The top/bottom 5–10% of several colon cancer cell lines with the highest/lowest fluorescence were sorted by FACS and designated the ALDHhigh/+ and ALDHlow/– cell population (Figure 2A). To compare the biological behaviour of these two sorted subpopulations, we observed their colony formation ability and growth curves in vitro. Compared to ALDHlow/– cells, ALDHhigh/+ cells (HCT116, SW480 and SW620 cells) generated more colonies after 14 days of incubation (Figure 2B) and grew faster (HCT116 and LOVO cells) (Figure 2C). We also examined the mRNA expression of 19 ALDH family members using qPCR in two groups sorted from six colon cancer cell lines, and found that the expression of ALDH1A1, ALDH1A3 and ALDH1L1 was significantly higher in ALDHhigh/+ cells than in counterpart cells (Figure 2D), and surprisingly, ALDH1A1 but not ALDH1A3 was the enzyme whose expression differed the most. Overall, ALDHhigh/+ colon cancer cells harbour highly clonogenic properties and aggressive growth in vitro, which is at least partially associated with ALDH1A3.
Isolation and characterisation of ALDHhigh/+ and ALDHlow/− colorectal cancer cells. (A) Flow diagram showing the gating of ALDHhigh/+(R1) and ALDHlow/−(R2) SW620 cells. (B) Colony formation assays after 14 days of incubation of 200 separated ALDHhigh/+ and ALDHlow/− colon cancer cells were compared. (C) The proliferation of isolated ALDHhigh/+ and ALDHlow/− HCT116 and LOVO cells were evaluated using MTT and CCK8 assays. (D) Real-time PCR of 19 ALDH isoforms in ALDHhigh/+ and ALDHlow/− cells from six colon cancer cell lines, each sample was analysed in triplicate and data shown was with the standard of CT SD⩽0.05, using the 2-ΔΔCT-method. The results shown are representative of at least three experiments; *P<0.05, **P<0.01, ***P<0.001.
ALDH1A3 siRNA reduced in vitro proliferation and invasion of HCT116 and A549 cells
The above data highlight that ALDH1A1 and ALDH1A3 might play important roles in ALDHhigh/+ cells, so we further examined whether altering the ALDH1A1 and ALDH1A3 expression levels could affect the cell proliferation and invasion. Since the relationship between ALDH1A3 and Aldefluor activity was especially close in pulmonary cancers and colorectal cancers (Figure 1D), the most widely used pulmonary cancer cell line (A549) and colon cancer cell line (HCT116) were chosen for siRNA experiments. Three pairs of ALDH1A3 siRNAs were transfected into HCT116 cells, and three pairs of ALDH1A1 siRNAs were transfected into A549 cells (ALDH1A1 was almost undetectable in HCT116 cells, and thus, all the ALDH1A1 siRNA experiments were performed in A549 cells). The knockdown effect was confirmed on both the mRNA and protein level (Figure 3A), and the most effective oligos (siALDH1A1-1 and siALDH1A3-3) were chosen for the subsequent experiments. Then, the effect of knockdown using siALDH1A1 and siALDH1A3 on ALDH enzyme activity was examined with Aldefluor assays. The percentage of ALDH+ HCT116 cells decreased from 67.2 to 17.5% 48 h after siALDH1A3-3 administration. For A549 cells, both siALDH1A1-1 and siALDH1A3-3 could effectively reduce the ALDH+ cell ratio (Figure 3B). The proliferation of HCT116 cells in vitro was reduced after transfection with siALDH1A3, and only siALDH1A3 but not siALDH1A1 attenuated the proliferation of A549 cells (Figure 3C). Cell cycle analysis of both HCT116 and A549 cells showed that the siALDH1A3 cell population contained more G0/G1 phase cells than the siNC cell population (Figure 3C). The in vitro invasion capability of both HCT116 and A549 cells was reduced after transfection with siALDH1A3 according to Transwell assays (Figure 3D). The two other siRNAs of ALDH1A3 (siALDH1A3-1 and siALDH1A3-2) were applied for proliferation assays in HCT116 and A549 cells to exclude off-target effects (Supplementary Figure 3). Collectively, knockdown of ALDH1A3 expression in HCT116 and A549 cells reduced their in vitro proliferation and invasion.
ALDH1A3 knockdown with siRNA reduced the propagation and invasion of HCT116 and A549 cells in vitro. (A) HCT116 and A549 cells were transfected with three pairs of siALDH1A3 and siALDH1A1, respectively. Real-time PCR (left) and western blotting (right) analysis showed the knockdown effectiveness. (B) siALDH1A3 in HCT116 cells, siALDH1A1 and siALDH1A3 in A549 cells reduced the ratio of ALDH+ cells, based on Aldefluor assays. (C) The proliferation of siALDH1A3 HCT116 cells and the proliferation of A549 cells after transfection with siALDH1A1 and siALDH1A3 was evaluated with CCK8 and MTT assays, respectively. (D) The in vitro invasion capability of HCT116 and A549 cells after transfection with siALDH1A3 was assayed using a Boyden-chamber assay. Values shown are for one representative experiment, the data are given as mean±s.d.; **P<0.01, ***P<0.001.
shRNA-mediated knockdown of ALDH1A3 in colon cancer cells has different effects on in vitro proliferation and invasion, perhaps due to contrasting effects on CXCR4 expression
To further evaluate the role of ALDH1A3 in malignancy, we established stable ALDH1A3-knockdown cells using two targeting shRNAs (sh-1A3-3 and sh-s30) in A549, HCT116, SW480, SW620 and LOVO cells. Both shRNAs could effectively reduce ALDH1A3 mRNA and protein expression (Figure 4C–E), and the number of ALDH+ cells was also dramatically reduced in the shALDH1A3 transfected cells (Supplementary Figure 4). To exclude off-target effects, mRNA expression of all the 19 ALDH family members in sh-control, sh-1A3-3 and sh-s30 cells was examined. The results showed that sh-s30 was more specific; thus, sh-s30 was used for further experiment (Supplementary Figure 5).
shRNA knockdown of ALDH1A3 in colon cancer cells had different effects due to the contrasting influence on CXCR4 expression. (A) The proliferation of sh-control (sh-scr) and sh-s30 cells was evaluated with CCK8 (SW480, HC116, LOVO and SW620) and MTT (A549) assays in vitro. (B) The in vitro invasion capability of sh-scr and sh-s30 cells was assayed using Transwell assays. (C) The respective mRNA level of a panel of 15 genes related to cell viability and migration was quantified by qPCR in sh-scr and sh-s30 SW480 cells. (D) Real-time PCR and (E) western blotting analysis showed that ALDH1A3 was downregulated in sh-1A3-3 and sh-s30 cells compared with the control sh-scr cells, while CXCR4 was downregulated in SW480, HC116 and A549 cells, upregulated in SW620 cells and exhibited no obvious change in LOVO cells. (F) The effect of CXCR4 overexpression in sh-s30 cells of SW480,A549 and LOVO cells was determined by qPCR. (G) The proliferation of sh-scr, sh-s30 and sh-s30+CXCR4 cells was evaluated with CCK8 assays in vitro. Experiments were repeated three times, values given are for one representative experiment. *P<0.05, **P<0.01, ***P<0.001.
Paradoxically, ALDH1A3 shRNA-knockdown resulted in different effect in different cell lines, that is, shALDH1A3-transduced HCT116, SW480 and A549 cells showed slower growth and less invasive capacity while downregulation of ALDH1A3 in SW620 and LOVO cells demonstrated no obvious proliferation changes (Figure 4A and B). To investigate how ALDH1A3 might work, we firstly performed an mRNA screen in a panel of 15 genes related to cell viability and migration in sh-control and sh-s30 SW480 cells. Among the various changes, an obvious decrease in CXCR4 expression was observed in shALDH1A3-transduced SW480 cells (Figure 4C). Then we examined the CXCR4 expression in all the shALDH1A3-cells, and found that CXCR4 change in sh-cells was consistent with their proliferation feature respectively. ALDH1A3 knockdown attenuated CXCR4 expression in HCT116, SW480 and A549 cells with a slower growth and less invasive capacity, but increased or did not significantly change CXCR4 expression in SW620 and LOVO, which showed no obvious phenotype changes (Figure 4A, B, D and E). This result might explain, at least partially, why shALDH1A3 in SW620 and LOVO did not reduce the cell proliferation. Then we transfected CXCR4 into sh-ALDH1A3 cells, and found that overexpression of CXCR4 could rescue cell growth suppression mediated by ALDH1A3 knockdown in SW480 and A549 cells, but had no effect on growth of LOVO cells (Figure 4F and G).
These observations suggest that knockdown of ALDH1A3 in colon cancer cells had a different influence on cell proliferation and invasion, which might rely on its differential effect on CXCR4, at least in the five cell lines used here.
ALDH1A3 expression is negatively correlated with CXCR4 in human cell lines
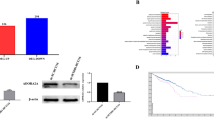
To further explore the possible relationship between ALDH1A3 and CXCR4, we qualified the mRNA expression of ALDH1A1, ALDH1A3, CXCR4 and CXCR7 using qPCR in the 58 human cell lines that had been used for the Aldefluor assay, and the results showed that ALDH1A3 and CXCR4 were negatively correlated (r=−0.387, P=0.003) (Figure 5A and B). The relative mRNA expression of CXCR4 was assayed by two pairs of primers, and the results were consistent (Supplementary Table 3). Furthermore, the protein expression of ALDH1A3 and CXCR4 in a panel of 63 human cell lines was examined, and the results also suggested a negative relationship between ALDH1A3 and CXCR4 (r=−0.34, P=0.01) (Figure 5C and D).
ALDH1A3 expression is inversely correlated with CXCR4 expression in human cell lines. (A) The mRNA expression of ALDH1A1, ALDH1A3, CXCR4 and CXCR7 in 58 human cell lines was quantified by qPCR, and the correlations between them were analysed with a Spearman rank correlation test, and the correlation coefficients (r) and statistically significant values (P) are shown. (B) The ALDH1A3 and CXCR4 mRNA expression in 58 human cell lines are displayed using an XY scatter plot. (C) Western blotting assays of ALDH1A3, CXCR4 and GAPDH in 63 human cell lines. (D) The correlation analysis between ALDH1A3 and CXCR4 protein levels is displayed in a scatter plot. The densitometric quantification was normalised to GAPDH expression, and SW480 was served as a control cell line.
Discussion
cancer stem cells, also called tumour-initiating cells, represent a small distinct subpopulation of tumour cells that are largely responsible for tumourigenesis and drug resistance. Compared with certain specific CSC markers, such as CD133, CD44, CD166, CD24 ABCG2 and EpCAM, ALDHs are regarded as universal functional markers and are widely used to isolate CSCs in various solid tumours, including breast cancer (Zhao et al, 2014), lung cancer (Shao et al, 2014), colon cancer (Huang et al, 2009), gastric carcinoma (Nguyen et al, 2016), prostate cancer (Cojoc et al, 2015) and melanoma (Luo et al, 2012). However, in human hepatocellular carcinoma, Tanaka found that ALDH1A1-overexpressing cells are differentiated cells rather than CSCs (Tanaka et al, 2015). In our study, the ALDH+ cell ratio was variable in the 58 human cell lines tested, but was consistently high in cancer cells from liver and kidney tissues (Supplementary Table 2). We also detected abundant expression of ALDH1A3 in 48 clear cell renal cell carcinomas (Supplementary Figure 6). This observation may be easily explained by the physical function of ALDH enzymes, but also brings up the question of whether ALDH could be used as a CSC marker in hepatocellular and renal cell carcinoma. Although this issue was not the focus of this study, the use of ALDH as a CSC marker is interesting and should be further investigated in future studies.
In our 58 human cell lines, the mRNA expression of ALDH1A3, rather than any other isotypes, was positively related to the Aldefluor activity. However, interestingly, in sorted ALDHhigh/+ colorectal cancer cells compared with ALDHlow/– counterparts, ALDH1A1 but not ALDH1A3 was the enzyme that showed the most predominant difference in expression, though ALDH1A3 was also higher in the ALDHhigh/+ subpopulation. This finding indicates a critical issue; the exact ratio of positive subtypes must be clarified when referring to ALDHhigh/+ cells.
To examine the function of ALDH1A3, we suppressed its expression in five human cell lines and observed an opposite influence on cell proliferation and invasion in different cells. Going forward, elucidating the biochemical mechanism of ALDH1A3 in cancer cell growth and survival will become critical. The ALDH1 family, including ALDH1A3, catalyses the conversion of retinaldehyde to RA in vitamin A metabolism. retinoic acid enters the nucleus and activates the RA receptors or the retinoid X receptors, which promote the expression of target gene that are involved in many important biological processes, including cell differentiation, proliferation and lipid metabolism. We screened the downstream genes of RA in parent and ALDH1A3-knockdown cells, and surprisingly, no meaningful change was found (Supplementary Figure 7). Unexpectedly, an obvious change in CXC chemokine receptor 4 (CXCR4) that accompanied ALDH1A3 knockdown was confirmed.
The CXCR4 belongs to the G protein-coupled receptor family, which influences various aspects of cancer cell biology, such as proliferation, migration and invasion, through a specific interaction with its ligand CXCL12 (stromal cell-derived factor-1, SDF-1) (Kucia et al, 2005). Recently, SDF-1 has been shown to bind to CXCR7, another member of the CXC chemokine receptor family (Heckmann et al, 2014). Few publications have elaborated on the relationship between CXCR4 and ALDH1A3. In our work, knocking down of ALDH1A3 induced divergent CXCR4 regulation in different colon cancer cells. One possible reason is there might be a negative feedback loop in ALDH enzyme family (Marcato et al, 2011b). Consistent with this knowledge, we found that there was usually an upregulation of some other ALDH isotypes accompanied by ALDH1A3 knockdown (Supplementary Figure 5). For example, mRNA expression of ALDH1B1, ALDH1L1 and ALDH16A1 increased in sh-ALDH1A3 SW480 cells, while abundant ALDH1A1 was detected in sh-ALDH1A3 HCT116 cells. Since different ALDH isoforms have a similar overlap in function, the final combined effect on CXCR4 could be an intricate integration. We also tested if all-trans retinoic acid which is produced by ALDH1A3 could similarly affect CXCR4 expression. The results showed all-trans retinoic acid treatment (10−7 M to 10−5 M) on SW480 and A549 cells for 24 h could not induce meaningful change of CXCR4 (Supplementary Figure 8), which suggested that other mechanism rather than the RA pathway might be responsible for the regulation of CXCR4 caused by sh-ALDH1A3.
To further validate the possible relationship of ALDH1A3 and CXCR4, we firstly examined their relative mRNA and protein expression in a large panel of 58 human cell lines regardless of the tissue origin and found a negative relationship. And then, we analysed the mRNA expression of ALDH1A3 and CXCR4 genes in 11 764 tumour samples from 37 different tissues of origin in TCGA (The Cancer Genome Atlas, http://xena.ucsc.edu/#analyze) (Hu et al, 2017). Interestingly, ALDH1A3 and CXCR4 were negatively related in thymoma (n=120, Spearman r=−0.52, P<0.001), but positively related in pancreatic adenocarcinoma (n=178, Spearman r =0.51, P<0.001). However, the relationship of those two genes in colon cancer was lower (n=457, Spearman r=0.33, P<0.001) (Supplementary Table 4 and Supplementary Figure 9). We propose that the controversial results might be due to ALDH1A3 playing different roles in different types of tissues, the complex relationship between ALDH1A3 and CXCR4 was tissue specific and sample specific. So until now, all we can say is ALDH1A3 and CXCR4 are interrelated, and the detailed mechanism how the two affect each other needs to be further explored.
In conclusion, we demonstrated that ALDH1A3 was the predominate ALDH isoform in human cell lines. The effect of ALDH1A3 on the malignant behaviour of tumour cells might be related to CXCR4 expression, but the detailed mechanism regarding how CXCR4 and ALDH1A3 interact with each other requires further exploration. This comprehensive study of ALDH isotypes will improve our understanding of the mechanisms of cancer, and the basic biological function of ALDH1A3 should be fully investigated further.
Change history
23 January 2018
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Canino C, Luo Y, Marcato P, Blandino G, Pass HI, Cioce M (2015) A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 6: 12637–12653.
Chen MH, Weng JJ, Cheng CT, Wu RC, Huang SC, Wu CE, Chung YH, Liu CY, Chang MH, Chiang KC, Yeh TS, Su Y, Yeh CN (2016) ALDH1A3, the major aldehyde dehydrogenase isoform in human cholangiocarcinoma cells, affects prognosis and gemcitabine resistance in cholangiocarcinoma patients. Clin Cancer Res 22: 4225–4235.
Cheng P, Wang J, Waghmare I, Sartini S, Coviello V, Kim S, Mohyeldin A, Pavlyukov MS, Zhang Z, Minata M, Valentim CL, Chhipa RR, Bhat KP, Dasgupta B, La Motta C, Kango-Singh M, Nakano I (2016) FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res 76: 7219–7230.
Cojoc M, Peitzsch C, Kurth I, Trautmann F, Kunz-Schughart LA, Telegeev GD, Stakhovsky EA, Walker JR, Simin K, Lyle S, Fuessel S, Erdmann K, Wirth MP, Krause M, Baumann M, Dubrovska A (2015) Aldehyde dehydrogenase is regulated by beta-catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res 75: 1482–1494.
Heckmann D, Maier P, Laufs S, Li L, Sleeman JP, Trunk MJ, Leupold JH, Wenz F, Zeller WJ, Fruehauf S, Allgayer H (2014) The disparate twins: a comparative study of CXCR4 and CXCR7 in SDF-1alpha-induced gene expression, invasion and chemosensitivity of colon cancer. Clin Cancer Res 20: 604–616.
Hu S, Yuan H, Li Z, Zhang J, Wu J, Chen Y, Shi Q, Ren W, Shao N, Ying X (2017) Transcriptional response profiles of paired tumor-normal samples offer novel perspectives in pan-cancer analysis. Oncotarget 8: 41334–41347.
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69: 3382–3389.
Kucia M, Reca R, Miekus K, Wanzeck J, Wojakowski W, Janowska-Wieczorek A, Ratajczak J, Ratajczak MZ (2005) Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF-1-CXCR4 axis. Stem Cells 23: 879–894.
Kurth I, Hein L, Mabert K, Peitzsch C, Koi L, Cojoc M, Kunz-Schughart L, Baumann M, Dubrovska A (2015) Cancer stem cell related markers of radioresistance in head and neck squamous cell carcinoma. Oncotarget 6: 34494–34509.
Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris DA, Roop DR, Vasiliou V, Fujita M (2012) ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells 30: 2100–2113.
Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I (2013) Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA 110: 8644–8649.
Marcato P, Dean CA, Giacomantonio CA, Lee PW (2011a) Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 10: 1378–1384.
Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, Wallace M, Clements D, Turner C, Mathenge EG, Gujar SA, Giacomantonio CA, Mackey JR, Godbout R, Lee PW (2015) Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol 9: 17–31.
Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, Giacomantonio CA, Lee PW (2011b) Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells 29: 32–45.
Marchitti SA, Brocker C, Stagos D, Vasiliou V (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4: 697–720.
Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD (2015) N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem Biol Interact 234: 18–28.
Nguyen PH, Giraud J, Chambonnier L, Dubus P, Wittkop L, Belleannee G, Collet D, Soubeyran I, Evrard S, Rousseau B, Senant-Dugot N, Megraud F, Mazurier F, Varon C (2016) Characterization of biomarkers of tumorigenic and chemoresistant cancer stem cells in human gastric carcinoma. Clin Cancer Res 23: 1586–1597.
Perez-Alea M, McGrail K, Sanchez-Redondo S, Ferrer B, Fournet G, Cortes J, Munoz E, Hernandez-Losa J, Tenbaum S, Martin G, Costello R, Ceylan I, Garcia-Patos V, Recio JA (2017) ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene 41: 5695–5708.
Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, Minna JD (2014) Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res 20: 4154–4166.
Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, Vasiliou V (2015) ALDH1B1 is crucial for colon tumorigenesis by modulating Wnt/beta-Catenin, notch and PI3K/Akt signaling pathways. PLoS One 10: e0121648.
Tanaka K, Tomita H, Hisamatsu K, Nakashima T, Hatano Y, Sasaki Y, Osada S, Tanaka T, Miyazaki T, Yoshida K, Hara A (2015) ALDH1A1-overexpressing cells are differentiated cells but not cancer stem or progenitor cells in human hepatocellular carcinoma. Oncotarget 6: 24722–24732.
Xu X, Chai S, Wang P, Zhang C, Yang Y, Wang K (2015) Aldehyde dehydrogenases and cancer stem cells. Cancer Lett 369: 50–57.
Yan J, De Melo J, Cutz JC, Aziz T, Tang D (2014) Aldehyde dehydrogenase 3A1 associates with prostate tumorigenesis. Br J Cancer 110: 2593–2603.
Zhang M, Liu Y, Feng H, Bian X, Zhao W, Yang Z, Gu B, Li Z (2013) CD133 affects the invasive ability of HCT116 cells by regulating TIMP-2. Am J Pathol 182: 565–576.
Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, Xiong Y, Guan KL, Lei QY (2014) NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Invest 124: 5453–5465.
Acknowledgements
We thank Dr Shuofeng Hu for correlation analysis of ALDH1A3 and CXCR4 genes in the TCGA data sets. This work has been supported by Beijing Natural Science Foundation (7154222), CAMS Innovation Fund for Medical Sciences (CIFMS) and National Science and Technology Infrastructure Grant NSTI-CR15/16.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Feng, H., Liu, Y., Bian, X. et al. ALDH1A3 affects colon cancer in vitro proliferation and invasion depending on CXCR4 status. Br J Cancer 118, 224–232 (2018). https://doi.org/10.1038/bjc.2017.363
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2017.363
Keywords
This article is cited by
-
Biological evaluation of levofloxacin and its thionated derivatives: antioxidant activity, aldehyde dehydrogenase enzyme inhibition, and cytotoxicity on A549 cell line
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Interrogating colorectal cancer metastasis to liver: a search for clinically viable compounds and mechanistic insights in colorectal cancer Patient Derived Organoids
Journal of Experimental & Clinical Cancer Research (2023)
-
Transcriptional responses of human intestinal epithelial HT-29 cells to spore-displayed p40 derived from Lacticaseibacillus rhamnosus GG
BMC Microbiology (2022)
-
Prognostic and clinicopathological insights of phosphodiesterase 9A gene as novel biomarker in human colorectal cancer
BMC Cancer (2021)
-
Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway
Oncogenesis (2021)