Abstract
Nuclear factor-κB (NF-κB) is an important regulator of cell survival and has been shown to be constitutively active in chronic lymphocytic leukemia (CLL) cells. Recently, a novel NF-κB inhibitor, IMD-0354 (N-(3, 5-bis-trifluoromethyl-phenyl)-5-chloro-2-hydroxy-benzamide), was shown to specifically inhibit the phosphorylation of IκBα by IkB kinases, thus preventing NF-κB release. In this study, we investigated if IMD-0354 can inhibit NF-κB activation and induce apoptosis in CLL cells in vitro. The rate of increase in apoptosis, drug sensitivity and DNA-binding activity of NF-κB were studied using Annexin V stainings, the fluorometric microculture cytotoxicity assay and electrophoretic mobility shift assay, respectively. Finally, the impact of IMD-0354 treatment on the expression of a set of apoptosis-related genes was investigated. The results clearly show that IMD-0354 induced apoptosis (mean 26%, range 8–48%) in CLL cells, independent of immunoglobulin heavy variable (IGHV) gene mutational status, and showed a dose-dependent cytotoxic effect. IMD-0354 treatment also significantly lowered the DNA-binding activity of NF-κB in CLL cells. In addition, we identified differences in expression levels of pro- and antiapoptotic genes following IMD-0354 treatment. In summary, our novel findings show that IMD-0354 can induce apoptosis in CLL cells, and thus merits further investigation as an anticancer agent in vivo.
Similar content being viewed by others
Main
During the past decade, major advances in the treatment of chronic lymphocytic leukemia (CLL) have been made, including the emergence of promising agents, such as the nucleoside analog fludarabine and the monoclonal antibodies alemtuzumab (anti-CD52) and rituximab (anti-CD20),1, 2 which have led to higher response rates and longer periods of remission for CLL patients. However, despite these therapeutic options, the disease will progress for many patients, for instance, in patients with poor-prognostic markers such as unmutated immunoglobulin heavy variable (IGHV) genes or deletions of 11q/17p. Hence, novel drugs for CLL treatment are needed, in which agents interfering with the nuclear factor-κB (NF-κB) pathway are one possible approach.3
NF-κB is an important regulator that not only mediates apoptotic signaling, but also plays critical roles in cell proliferation, cell adhesion, inflammation, differentiation, angiogenesis and tumor cell invasion.4, 5 Several studies suggest that CLL cells display constitutive activation of NF-κB, which results in a block of apoptosis for the malignant B cells.3, 6, 7 NF-κB is a collective name for inducible dimeric transcription factors composed of members of the Rel family of DNA-binding proteins that recognize a common DNA sequence motif. Human beings express five NF-κB/Rel proteins: p50, p52, p65 (RelA), c-Rel and RelB, in which homo/heterodimers containing different combinations of these subunits are held in the cytoplasm through interaction with specific inhibitors, the IkBs (IkBα, IkBβ and IkBγ). The process leading to the activation of NF-κB requires phosphorylation of IkB by IkB kinases (IKK), resulting in the degradation of IkB by rapid ubiquitin-dependent degradation. This allows the translocation of NF-κB from the cytosol to the nucleus, in which the heterodimer binds to a response element in the promoter region of specific target genes. Previous studies have indicated that NF-κB inhibitors, such as LC-1 and DHMEQ, can induce apoptosis in CLL cells, especially when combined with fludarabine.8, 9 These inhibitors have also been able to accelerate apoptosis in CLL cells.10 Deguelin is another drug that has been shown to inhibit IkBα in CLL cells, also leading to the induction of apoptosis.11
Recently, the compound IMD-0354 (N-(3, 5-bis-trifluoromethyl-phenyl)-5-chloro-2-hydroxy-benzamide), which is a novel inhibitor of IkB phosphorylation, was shown to induce apoptosis in human mast cells with constitutively activated c-kit receptors and was suggested to have therapeutic activity.12 Specifically, IMD-0354 inhibits the IKK-induced phosphorylation of IkBα and hence protects it from undergoing degradation, which in turn results in the inhibition of NF-κB activation. In this study, we investigated the effect of IMD-0354 in CLL cells in vitro by performing Annexin V stainings and a non-clonogenic cytotoxicity assay. Furthermore, we investigated the effect of IMD-0354 treatment on gene expression of different pro- and antiapoptotic genes. Our data clearly show that IMD-3054 can induce apoptosis of CLL cells directly by inhibiting the NF-κB pathway and should be further investigated as a new potential drug in CLL treatment.
Materials and methods
Patient samples
In total, tumor samples from 20 untreated CLL patients were included in this study. CLL cells displayed the typical immunophenotype and met the recently revised diagnostic criteria.13 Tumor material was obtained from peripheral blood samples. The median age at diagnosis was 64 years (range, 44–85 years), with a male : female ratio of 4 : 1. Eleven cases displayed IGHV unmutated genes and eight cases IGHV mutated genes, whereas in one case the mutation status was unknown. Informed consent was obtained according to the Declaration of Helsinki and ethically approved by the local ethical review committee.
Cell culture, drug treatment and Annexin V stainings
CLL cells were cultured in RPMI 1640 medium with 10% fetal calf serum (Gibco, Paisley, UK) supplemented with the addition of 4 mM glutamine and 1 × penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA) at a cell density of 1 × 106 cells per ml at 37 °C in 5% CO2 atmosphere. CLL cell cultures were maintained for 48 h both with/without the addition of IMD-0354 (Sigma Aldrich, St Louis, MO, USA) 1–10 μM and the survival factor thioredoxin (Trx)14 (1 μg/ml) (Sigma Aldrich). Annexin V staining for detection of phosphatidyl serine exposed on the plasma membrane was carried out using Annexin V-fluorescein isothiocyanate apoptosis detection kit (BD Biosciences Pharmingen, San Diego, CA, USA) according to the manufacturer's instruction. In brief, cells were washed and incubated for 15 min in the dark with ITC-conjugated Annexin V and propidium iodide provided with the kit and a minimum of 10 000 events were collected on a Beckman Coulter FACSort flow cytometer. Apoptosis was calculated as the percentage of Annexin V-positive cells corrected for the percentage occurring owing to spontaneous apoptosis in control wells.
Fluorometric microculture cytotoxicity assay
The fluorometric microculture cytotoxicity assay is a total cell kill assay, based on the ability of cells with intact cell membranes to convert non-fluorescent diacetate to fluorescent fluorescein.15 Tumor cells from eight CLL samples were cultured in 384-well microtiter plates that contained different concentrations of IMD-0354 ranging from 0.01 to 100 μM, and all samples were run in duplicates. The plates were incubated at 37 °C and 5% CO2 for 72 h and then analyzed using the automated Optimized Robot for Chemical Analysis (Orca, Beckman Coulter, Fullerton, CA, USA). Both the culture medium and drug were aspirated from the plates, followed by two rounds of cell washes in phosphate-buffered saline. Q2 buffer (containing 125 mM NaCl and 25 mM Hepes buffer, pH 7.4) was added before the addition of 10 μg/ml of fluorescein diacetate (Sigma Aldrich) to each well. The plates were incubated for 50 min and fluorescence was measured in Fluostar Optima (BMG Technologies, Offenburg, Germany) at 485 nm/520 nm. Cell survival after exposure to the cytotoxic drug was presented as the percentage of survival index, defined as mean fluorescence in duplicate test wells in the percentage of control wells with blank values subtracted. Thus, a low numerical value indicates a high cytotoxic effect. A successful assay required a ratio of >5 between the signal in the control wells and the blank wells, a coefficient of variation of <30% in the control wells and a tumor cell fraction exceeding 70% in the cell preparation.
Multiplex ligation-dependent probe amplification analysis
Cells from five CLL cases were cultured in the presence of the tumor necrosis factor-α (25 ng/ml), which was used to activate the NF-κB pathway, for 24 h with the subsequent the addition of IMD-0354 (1 μM) and collected after an additional 24 h. RNA was purified using the Ambion RNA extraction kit (Applied Biosystems, Foster City, CA, USA) and concentrations measured utilizing the NanoDrop instrument. Quality of RNA was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Multiple ligation-dependent probe amplification (MLPA) RNA analysis was carried out using SALSA RT-MLPA KIT R011-B1 Apoptosis mRNA (MRC-Holland, Amsterdam, the Netherlands), for the analysis of 35 apoptosis-related genes as per the manufacturer's instruction. Briefly, 100 ng of total RNA was reverse transcribed using the RT primer mix provided by the kit, which contains one RT primer for each of the 35 different apoptosis-related genes and four control genes in a single multiplex reaction. MLPA probes were subsequently added and allowed to hybridize for 16 h at 60 °C. Following hybridization, the probes were ligated with ligase 65 and polymerase chain reaction amplified utilizing FAM-labeled M13 primers. All polymerase chain reaction products were analyzed on the Applied Biosystems 9700 DNA sequencer (Applied Biosystems) using the GeneScan software. Peak tables for each sample were exported from GeneScan to Microsoft Excel, in which data normalization to the control gene β-2-microglobulin was carried out. Normalized data sets were grouped and analyzed for significant differences to the Statistica software (ver.7.1, Stat Soft, Tulsa, OK, USA) using the χ2-test (significance P<0.05). A heat map for visualization purposes was generated in the open source multiexperiment viewer software (http://www.tm4.org/mev), in which data were adjusted to a 0–1 scale for color expression coding.
NF-κB activation by electrophoretic mobility shift assay
CLL cells (1 × 106 cells per ml) were treated both with/without 1 μM of IMD-0354 for 24 h. Cytoplasmic and nuclear extracts were prepared using hypotonic buffer (20 mm Hepes, pH 7.6, 20% (w/v) glycerol, 20 mm NaCl, 10 mm MgCl2, 0.2 mm EDTA, 1% (w/v) Triton X-100, 1 mm dithiothreitol, 1 mm sodium orthovanadate and 1% aprotinin) and hypertonic buffer (20 mm Hepes, pH 7.6, 20% (w/v) glycerol, 500 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1% (w/v) Triton X-100, 1 mm dithiothreitol, 1 mm sodium orthovanadate and 1% aprotinin), respectively. Briefly, cells were washed twice with phosphate-buffered saline, and lysed in hypotonic buffer, for 10 min on ice. Nuclei were spun down and incubated in hypertonic lysis buffer for 20 min on ice. Supernatants were removed and protein concentrations were determined. The NF-κB-specific consensus oligonucleotide (5′-AGTTGAGGGTTTCCCAGGC-3′; Promega, Madison, WI, USA) was 5′ end labeled with [γ-32P]adenosine triphosphate using polynucleotide kinase (Gibco, Gaithersburg, MD, USA). For the binding reactions, equal quantities of nuclear extracts (1–4 μg) were incubated with 10 000 c.p.m. of end-labeled NF-κB oligonucleotide and 1 μg of poly dI–dC (GE Health Care, Buckinghamshire, UK) in the binding buffer (20 mm Hepes, pH 7.6, 20% (w/v) glycerol, 100 mm NaCl, 10 mm MgCl2, 0.2 mm EDTA, 1% (w/v) Triton X-100, 1 mm dithiothreitol). Free oligonucleotide and oligonucleotide-bound proteins were separated by electrophoresis on a native 6% polyacrylamide gel. Gels were dried under vacuum on Whatman paper and exposed to Kodak BioMax MS film (Sigma Aldrich) for 3–6 h at −80 °C. The absence of binding in the presence of excess unlabeled NF-κB-specific oligonucleotide (competitor oligonucleotide) confirmed NF-κB-binding specificity.
Results
IMD-0354 induces apoptosis in CLL cells in vitro
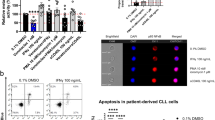
To determine whether IMD-0354 is capable of inducing apoptosis in CLL cells in vitro, cells from 20 CLL patients were incubated in the presence and absence of IMD-0354. The survival factor Trx was added in cells from 10 of these patients (five IGHV mutated and five IGHV unmutated samples). The percentage of early apoptotic and viable cells was evaluated by Annexin V/IP flow cytometry after 48 h of incubation. The Annexin V-positive percentage of cells was calculated as an increase over spontaneous apoptosis in untreated controls samples at that time point. Overall, the majority of CLL samples investigated showed an increase in the percentage of apoptotic cells following treatment with IMD-0354 only (mean 26%, range 8–48%), whereas treatment with IMD-0354+Trx gave a somewhat lower mean percentage of apoptosis (mean 20%, range 10–30%) (Figures 1a and b). On the contrary, CLL cells treated with Trx alone (n=10) did not show any apoptosis. Out of total 20 CLL samples with IMD-0354 treatment, the percentage of average mean increase in apoptosis for all unmutated samples was 30% and for all mutated samples 25%.
IMD-0354 induces apoptosis in CLL cells in vitro. (a) The percentage of increased apoptosis in CLL samples (n=20) cultured in vitro with 1 μM IMD-0354 either alone (n=10) or in the presence of Trx (n=10). (b) Representative flow cytometry analysis of apoptosis in CLL cells with and without IMD-0354 treatment after 48 h of incubation. (c) Dose response to IMD-0354 after incubation for 24 h, as measured by Annexin V, based on the average results after culturing two CLL samples. All cultures were carried out in duplicate.
CLL cells in duplicate (from two patients) were also incubated with increasing amounts of IMD-0354, with concentrations ranging from 1 to 10 μM. A dose-dependent increase in the apoptotic response of CLL cells was observed after 24 h of IMD-0354 incubation (Figure 1c).
Dose-dependent inhibition of CLL cell survival in vitro
To measure the cytotoxicity of IMD-0354 and calculate the percentage of surviving CLL cells, we performed fluorometric microculture cytotoxicity assay. Tumor cells from eight CLL samples were cultured in 384-well microtiter plates with different concentrations of IMD-0354. The percentage of cell survival was calculated as outlined in the Materials and methods section. As shown in Figure 2, all CLL patient samples showed a dose-dependent decrease in the percentage of survival index, with increasing concentrations of IMD-0354.
Inhibition of NF-κB activation in CLL cells
To determine the DNA-binding activity of NF-κB in both IMD-0354-treated and -untreated CLL cells, for four CLL samples, we performed electrophoretic mobility shift assay, using a small consensus NF-κB binding site. As shown in Figure 3, the DNA-binding activity of NF-κB is almost lost in the IMD-0354-treated CLL cells (lanes 6–9), as compared with the untreated cells, which displayed a high DNA-binding activity (lanes 2–5). The cold competition experiment, which was performed using 100-fold excess of unlabeled oligonucleotide, showed a complete loss of DNA-binding activity (lane 10). These results indicate that the activity of NF-κB is significantly lowered following 24 h of IMD-0354 treatment in all four CLL samples.
Electrophoretic mobility shift assay with an NF-κB consensus oligonucleotide using nuclear extracts from four CLL samples with and without IMD-0354 treatment. Lane 1, probe alone; lanes 2–5, CLL patient samples without IMD-0354 treatment; and lanes 6–9, CLL samples treated with 1 μM IMD-0354. In lane 10, a 100-fold excess of unlabeled probe was added to the reaction, containing the same lysates from CLL patient 1 sample mix (lane 2) without IMD-0354 treatment.
Induction of apoptotic-related gene expression in CLL cells
The above results indicated that IMD-0354 significantly induced apoptosis by targeting NF-κB-inducing genes and hence decreased the cell survival in CLL patient samples in vitro. To test for changes in the expression of apoptotic genes in both IMD-0354-treated and -untreated CLL cells, the expression changes of 35 different apoptosis-related genes was investigated by applying the RT-MLPA kit in five CLL samples. Cells were first treated with tumor necrosis factor-α for 24 h, with subsequent addition of IMD-0354 for another 24 h and thereafter total RNA was extracted. Significant differences (P<0.05) in gene expression were only observed for three genes (BIRC2, BIRC3 and BBC3) (Figure 4). To confirm gene expression levels observed with MLPA, we selected four genes (BCL2, BCL2A1, BAX and MCL2) and verified the results using quantitative real-time-polymerase chain reaction (data not shown).
Multiplex ligation-dependent probe amplification analysis before and after IMD-0354 treatment. (a) Apoptotic-related MLPA expression data for five CLL samples without (column 1–5) or with (column 6–10) IMD-0354 treatment. All the genes are listed on the right side of the image. Red color indicates increased and green color decreased gene expression. (b) The relative expression levels of apoptotic-related genes with significant differences (P<0.05) before and after IMD-0354 treatment.
Discussion
Many studies have shown that constitutive activation of NF-κB is a hallmark for most types of human cancer including leukemia and lymphoma.16, 17 Given the considerable contribution of NF-κB to proliferation in many cancers, NF-κB could be one of the important molecular targets of therapy and inhibition of NF-κB may also have a therapeutic potential for the control of tumor progression. In this study, we applied a novel drug, IMD-0354, to inhibit NF-κB activation in CLL cells in vitro. IMD-0354 has been shown to inhibit proliferation of neoplastic cells and results in suppressed progression in human breast cancer and mast cells.12, 18 It has also been shown in mice models that IMD-0354 treatment can significantly reduce colitis-associated tumorogenesis.19 However, to our knowledge, the effect of IMD-0354 in CLL has previously not been investigated.
The phenotype of CLL cells is characterized by defects in both programmed cell death and control of proliferation.20 When applying IMD-0354, a clear increase in apoptosis was observed in the majority of CLL samples investigated (Figure 1a). As it is well known that CLL cells will rapidly undergo spontaneous apoptosis when cultured in vitro,21 we also added a survival factor (Trx)14 when performing the IMD-0354 experiments, to prevent or reduce apoptosis in CLL cells. Despite this, a similar increase in the level of apoptosis was observed for treatments, including IMD-0354 alone or IMD-0354 along with Trx, which indicates that IMD-0354 indeed can induce apoptosis in CLL cells in vitro (Figure 1). Importantly, IMD-0354 also induced apoptosis at similar levels in both IGHV mutated and unmutated subgroups of CLL cases, indicating that both these prognostic subgroups respond similarly to the effect of IMD-0354. In addition, we measured the cytotoxic effect of IMD-0354 on the CLL cells using fluorometric microculture cytotoxicity assay, and all cases showed an increase in cytotoxicity in a dose-dependent manner (Figure 2).
IMD-0354 is thought to induce apoptosis by inhibiting the degradation of IkB and thereby protecting it against phosphorylation, which in turn inhibits NF-κB activation. To show the direct inhibition of NF-κB activity, we performed electrophoretic mobility shift assay to determine and compare the DNA-binding activity of NF-κB before and after the IMD-0354 treatment in CLL cell nuclear extracts. Significant reduction in DNA-binding activity of NF-κB was observed after IMD-0354 treatment (Figure 3), hence indicating that IMD-0354 indeed induced apoptosis in CLL cells by direct targeting of NF-κB.
Reduction of NF-κB may result in significant changes in expression patterns of apoptotic-related genes, which may include both anti- and proapoptotic BCL2 family members. The BCL2 proteins are well-characterized regulators of apoptosis and can both promote survival (antiapoptotic) and induce cell death (proapoptotic), and hence play an important role in oncogenesis.22, 23 We explored possible changes in gene expression of several apoptosis-related genes before and after the IMD-0354 treatment in CLL cells using the MLPA technique. After the IMD-0354 treatment, significant reduction of gene expression was observed for the antiapoptotic BIRC3 and BIRC2 genes. These two genes are IAP family members, which are known to be highly expressed in cancer cells compared with normal cells.24 BBC3, also known as PUMA, was the only proapoptotic gene that showed significant increase in the expression after IMD-0354 treatment. PUMA is a proapoptotic member of the BCL2 family, which mediates p53-dependent and -independent apoptosis.25 Hence, these results suggest that IMD-0354 induces apoptosis in CLL cells by decreasing and increasing anti- and proapoptotic genes, respectively.
Taken together, we conclude that IMD-0354 specifically targets the NF-κB pathway, and significantly induces apoptosis in CLL cells in vitro. This effect was retained when adding the survival factor Trx and the drug was equally effective in subgroups defined by the IGHV mutational status. We encourage further investigations of this drug as it may represent a novel potential drug in CLL treatment.
References
Hallek M . State-of-the-art treatment of chronic lymphocytic leukemia: a review. Hematol Am Soc Hematol Educ Program 2009, 440–448.
Ricci F, Tedeschi A, Morra E, Montillo M . Fludarabine in the treatment of chronic lymphocytic leukemia: a review. Ther Clin Risk Manag 2009; 5: 187–207.
Lopez-Guerra M, Colomer D . NF-kappaB as a therapeutic target in chronic lymphocytic leukemia. Expert Opin Ther Targets 2010; 14: 275–288.
Rahman KM, Li Y, Sarkar FH . Inactivation of akt and NF-kappaB play important roles during indole-3-carbinol-induced apoptosis in breast cancer cells. Nutr Cancer 2004; 48: 84–94.
Rahman KW, Sarkar FH . Inhibition of nuclear translocation of nuclear factor-{kappa}B contributes to 3,3′-diindolylmethane-induced apoptosis in breast cancer cells. Cancer Res 2005; 65: 364–371.
Cuni S, Perez-Aciego P, Perez-Chacon G, Vargas JA, Sanchez A, Martin-Saavedra FM et al. A sustained activation of PI3K/NF-kappaB pathway is critical for the survival of chronic lymphocytic leukemia B cells. Leukemia 2004; 18: 1391–1400.
Furman RR, Asgary Z, Mascarenhas JO, Liou HC, Schattner EJ . Modulation of NF-kappa B activity and apoptosis in chronic lymphocytic leukemia B cells. J Immunol 2000; 164: 2200–2206.
Hewamana S, Lin TT, Jenkins C, Burnett AK, Jordan CT, Fegan C et al. The novel nuclear factor-kappaB inhibitor LC-1 is equipotent in poor prognostic subsets of chronic lymphocytic leukemia and shows strong synergy with fludarabine. Clin Cancer Res 2008; 14: 8102–8111.
Horie R, Watanabe M, Okamura T, Taira M, Shoda M, Motoji T et al. DHMEQ, a new NF-kappaB inhibitor, induces apoptosis and enhances fludarabine effects on chronic lymphocytic leukemia cells. Leukemia 2006; 20: 800–806.
Pickering BM, de Mel S, Lee M, Howell M, Habens F, Dallman CL et al. Pharmacological inhibitors of NF-kappaB accelerate apoptosis in chronic lymphocytic leukaemia cells. Oncogene 2007; 26: 1166–1177.
Geeraerts B, Vanhoecke B, Vanden Berghe W, Philippe J, Offner F, Deforce D . Deguelin inhibits expression of IkappaBalpha protein and induces apoptosis of B-CLL cells in vitro. Leukemia 2007; 21: 1610–1618.
Tanaka A, Konno M, Muto S, Kambe N, Morii E, Nakahata T et al. A novel NF-kappaB inhibitor, IMD-0354, suppresses neoplastic proliferation of human mast cells with constitutively activated c-kit receptors. Blood 2005; 105: 2324–2331.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute—Working Group 1996 guidelines. Blood 2008; 111: 5446–5456.
Nilsson J, Soderberg O, Nilsson K, Rosen A . Thioredoxin prolongs survival of B-type chronic lymphocytic leukemia cells. Blood 2000; 95: 1420–1426.
Nygren P, Fridborg H, Csoka K, Sundstrom C, de la Torre M, Kristensen J et al. Detection of tumor-specific cytotoxic drug activity in vitro using the fluorometric microculture cytotoxicity assay and primary cultures of tumor cells from patients. Int J Cancer 1994; 56: 715–720.
Karin M, Cao Y, Greten FR, Li ZW . NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002; 2: 301–310.
Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet Jr RJ, Sledge Jr GW . Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol Cell Biol 1997; 17: 3629–3639.
Tanaka A, Muto S, Konno M, Itai A, Matsuda H . A new IkappaB kinase beta inhibitor prevents human breast cancer progression through negative regulation of cell cycle transition. Cancer Res 2006; 66: 419–426.
Hayakawa Y, Maeda S, Nakagawa H, Hikiba Y, Shibata W, Sakamoto K et al. Effectiveness of IkappaB kinase inhibitors in murine colitis-associated tumorigenesis. J Gastroenterol 2009; 44: 935–943.
Chiorazzi N . Cell proliferation and death: forgotten features of chronic lymphocytic leukemia B cells. Best Pract Res Clin Haematol 2007; 20: 399–413.
Collins RJ, Verschuer LA, Harmon BV, Prentice RL, Pope JH, Kerr JF . Spontaneous programmed death (apoptosis) of B-chronic lymphocytic leukaemia cells following their culture in vitro. Br J Haematol 1989; 71: 343–350.
Danial NN, Korsmeyer SJ . Cell death: critical control points. Cell 2004; 116: 205–219.
Reed JC . Apoptosis mechanisms: implications for cancer drug discovery. Oncology (Williston Park) 2004; 18 (Suppl 10): 11–20.
Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet 2003; 12: 791–801.
Kim MR, Jeong EG, Chae B, Lee JW, Soung YH, Nam SW et al. Pro-apoptotic PUMA and anti-apoptotic phospho-BAD are highly expressed in colorectal carcinomas. Dig Dis Sci 2007; 52: 2751–2756.
Acknowledgements
This study was supported by grants from the Swedish Cancer Society, the Swedish Society for Medical Research and Lion's Cancer Research Foundation, Uppsala. We thank Elin Lindhagen and Ida Lindbom for their great support during the study. MK and GT performed research, analyzed data and wrote the paper; AÅ provided samples and associated data; KN and RR designed and supervised the research and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
At the time of the study Anna Åleskog worked as a hematologist at the Department of Hematology, Uppsala University Hospital, but is currently employed as medical affairs manager at MSD, Sweden. Gerard Tobin worked at the time of the study at Rudbeck Laboratory, Uppsala University, Sweden, but is currently employed by Hofmann La Roche, Switzerland.
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kanduri, M., Tobin, G., Åleskog, A. et al. The novel NF-κB inhibitor IMD-0354 induces apoptosis in chronic lymphocytic leukemia. Blood Cancer Journal 1, e12 (2011). https://doi.org/10.1038/bcj.2011.9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2011.9
Keywords
This article is cited by
-
Inhibition of kinase IKKβ suppresses cellular abnormalities induced by the human papillomavirus oncoprotein HPV 18E6
Scientific Reports (2021)
-
Numblike regulates proliferation, apoptosis, and invasion of lung cancer cell
Tumor Biology (2013)
-
Evaluation of chronic lymphocytic leukemia by oligonucleotide-based microarray analysis uncovers novel aberrations not detected by FISH or cytogenetic analysis
Molecular Cytogenetics (2011)