Abstract
The t(11;14)(q13;q32) resulting in cyclin D1 overexpression is consistently present in mantle cell lymphoma. However secondary chromosomal aberrations are also extremely common. Of these, 8q24 abnormalities associated with the t(11;14) are rare. Over the course of 10 years at M.D. Anderson Cancer Center, we identified five cases of mantle cell lymphoma in which conventional cytogenetic analysis revealed complex karyotypes, including the t(11;14) and 8q24 abnormalities: one with t(8;14)(q24;q32), one with t(2;8)(q13;q24), and three with add(8)(q24). We performed fluorescence in situ hybridization (FISH) studies on all cases. In the case with the t(8;14), IgH/myc fusion signals were identified, and in the case with the t(2;8), split c-myc signals were detected. In the three cases with add(8)(q24), one case had split c-myc signals and two cases had three copies of c-myc. Thus, the c-myc gene was involved in all cases. All five neoplasms had blastoid morphologic features, and four cases, including the cases with the t(8;14) and t(2;8), had leukemic involvement. We conclude that 8q24 abnormalities involving the c-myc gene are uncommon secondary abnormalities that occur in a subset of mantle cell lymphomas. C-myc gene abnormalities are associated with blastoid cytologic features and also may be associated with leukemic involvement.
Similar content being viewed by others
INTRODUCTION
Mantle cell lymphoma is a type of B-cell non-Hodgkin’s lymphoma with distinctive histologic and immunophenotypic findings, and virtually all cases carry the t(11;14)(q13;q32) (1). In this translocation, the ccnd-1 gene at chromosome 11q13 is juxtaposed with the IgH gene on chromosome 14q32, resulting in overexpression of cyclin D1 (1, 2, 3). Although the t(11;14) is thought to be important in the pathogenesis of mantle cell lymphoma, this translocation is not sufficient to induce lymphomagenesis in transgenic mice experiments (4, 5), suggesting that other abnormalities are involved. Furthermore, conventional cytogenetic studies have shown that the t(11;14) is rarely an isolated finding in mantle cell lymphoma. Multiple other karyotypic aberrations are frequent, suggesting the importance of additional molecular abnormalities in the pathogenesis of these neoplasms (6, 7, 8, 9, 10, 11, 12, 13). Of these, a small number of mantle cell lymphomas with 8q24 abnormalities have been reported in the literature (14, 15, 16, 17, 18, 19).
We report five additional cases of mantle cell lymphoma in which conventional cytogenetic analysis showed complex karyotypes including the t(11;14) and abnormalities involving the 8q24 locus. All cases had blastoid morphologic features, and four patients had leukemic involvement. We performed fluorescence in situ hybridization (FISH) analysis and showed involvement of the c-myc gene in all cases.
MATERIALS AND METHODS
Case Selection
Five cases of mantle cell lymphoma in which conventional cytogenetic studies demonstrated the t(11;14)(q13;q32) and complex chromosomal abnormalities, including aberrations of the 8q24 locus, were obtained through a search of the files from 1991–2001 of the Department of Hematopathology at the University of Texas M.D. Anderson Cancer Center. Four of these cases were briefly mentioned as part of a cytogenetic study of 46 cases of mantle cell lymphoma reported previously (13).
Clinical information was obtained by review of the medical record. Wright-Giemsa–stained peripheral blood and bone marrow aspirate smears and hematoxylin-eosin–stained histological sections of bone marrow aspirate clot, core biopsy and lymph node, or other tissue biopsy specimens were reviewed. Complete blood counts and differential counts were performed for all patients, but the differential count at time of initial diagnosis was not available for Case 5. Each patient underwent one or more bone marrow aspiration and biopsy procedures during the course of disease.
Immunophenotypic Methods
Bone marrow aspirate, peripheral blood, or cell suspensions of tissue biopsy specimens of all five cases were assessed using three-color flow cytometry analysis and a FACScan (Becton Dickinson, San Jose, CA) instrument as described previously (20). Lymphocytes were gated for analysis using CD45 expression and side scatter, as described by Borowitz and colleagues (21). The CD45 antibody used for gating was conjugated to peridin chlorophyll alpha protein. Fluorescein isothiocyanate (FITC)- and phycoerythrin (PE)-conjugated antibodies were used as negative controls, and cursors were set to include >95% of events as negative. The panel of antibodies included reagents specific for CD3, CD4, CD5, CD7, CD10, CD19, CD20, CD23, FMC-7, and immunoglobulin kappa and lambda light chains (Becton-Dickinson).
Immunohistochemical studies were performed using formalin-fixed, paraffin-embedded tissue sections of bone marrow aspirate clot, core biopsy, and/or lymph node biopsy specimens. Cyclin D1 was assessed immunohistochemically in all cases. After deparaffinization and dehydration in graded alcohols and xylene, endogenous peroxidase was blocked with hydrogen peroxide. Heat-induced epitope retrieval was performed using citrate buffer, pH 6.0. After rinsing in phosphate-buffered saline, mouse anti-cyclin D1 antibody (DCS-6; DAKO, Carpinteria, CA) was applied at a dilution of 1:350. The LSAB2 kit (DAKO) was used as the detection system. Negative controls were also run. Other antibodies specific for CD3, CD5, CD10, CD20, CD23, CD30, CD43, BCL-2, Ki-67, and terminal deoxynucleotidyl transferase (TdT) were used in selected cases during the workup to establish the diagnosis.
Conventional Cytogenetic Studies
Conventional G-band karyotype analysis was performed on bone marrow aspirate specimens of all cases, as described previously (20). Cells were placed in 10 mL of Ham F10 medium, with 20% fetal serum at a concentration of 2 × 106 to 4 × 106 nucleated cells per milliliter. The culture was incubated at 37° C for approximately 24 hours. Standard harvesting procedures were used. Colcemid (0.1 mL/10 mL) was added to the culture for 30 minutes at room temperature. For hypotonic treatment, 0.075 mol/L KCl was used for 30 minutes at room temperature. The fixation procedure consisted of three changes of methanol/glacial acetic acid (3:1), with a 10-minute interval between each change. A drying chamber (Thermaton Industries, Holland, MI) was used for slide preparation. Slides were placed in a 60° C oven overnight, followed by GTG banding. The karyotype reports were written using the International System for Human Cytogenetic Nomenclature (22).
Fluorescence In Situ Hybridization Methods
The LSI IGH/MYC/CEP8 dual-fusion translocation probe and LSI MYC dual color break-apart probes were used in this study. Both probes are commercially available (Vysis, Downers Grove, IL). The FISH assay was performed using 5-μm-thick tissue sections of formalin-fixed, paraffin-embedded bone marrow biopsy specimens. The tissue sections were deparaffinized in Hemo-De (3 cycles, at ambient temperature), dehydrated in 100% ethanol (room temperature), and air-dried by placing the slides on a 45–50° C slide warmer for 2–5 minutes. The slides were then immersed in 0.2 N HCL for 20 minutes, followed by washes (3 minutes each) in purified water and wash buffer (2 × SSC, pH 7.0). Pretreatment consisted of immersion in pretreatment solution (sodium thiocyanate, Vysis) at 80° C for 30 minutes, again followed by washes in purified water and wash buffer. The tissue sections were digested in protease solution at 37° C for 10 minutes, washed in buffer, and dried on a slide warmer for 2–5 minutes. The slides were fixed in 10% formalin/PBS for 10 minutes (at ambient temperature) and washed in buffer solution. Co-denaturation of the probe mixture and specimen was performed at 80° C (5 minutes) using the Vysis HYBrite hybridization system. The probe mixture included 7 μL of LSI hybridization buffer (Vysis), 1 μL of probe, and 2 μL of purified water. After an overnight hybridization at 37° C, the slides were washed in 2 × SCC/0.1% NP-40 at 73° C for 2 minutes, rinsed in 2 × SSC/0.1NP-40 for 5–10 seconds at room temperature, air dried in darkness, and counterstained with 10 μL of 4, 6-diamidino-2-phenylindole in antifade (DAPI).
Hybridization signals were analyzed using an Olympus BX51 microscope equipped with appropriate filters and images and captured using the Cyto Vision imaging system (Applied Imaging, Santa Clara, CA).
RESULTS
Clinical Data
The clinical data are summarized in Table 1. All five patients were referred to this institution after initial diagnosis at another hospital. There were three women and two men, with a median age of 60 years and a mean age of 62.6 years (range, 50–74 y). Three patients had lymphadenopathy and underwent lymph node biopsy (Cases 1, 2, and 5). Three patients had splenomegaly (Cases 1, 2, and 4), and one patient had histologically proven gastrointestinal tract involvement (Case 5). All patients had an elevated serum lactate dehydrogenase (LDH) level. Three patients had serum LDH levels of 692, 1046, and 1826 IU/L (normal range, 313–618 IU/L). Two other patients (Cases 2 and 4) also had an elevated serum LDH level by history determined at another hospital, but the numerical value was not available.
All patients had an increased white blood cell count with absolute lymphocytosis at the time of initial diagnosis. The absolute lymphocyte count is available in four patients and ranged from 13.6 to 392.9 × 109/L (13.6–392.9/mm3; median, 137.8 × 109L [137.8 per mm3]). The percentage of lymphocytes was not available for one patient (Case 5). Three patients presented during the initial workup with an overt leukemic phase with a markedly elevated white blood cell count and absolute lymphocytosis. Two patients (Cases 4 and 5) had a modestly increased white blood cell count and modest absolute lymphocytosis at time of initial examination. Flow cytometry immunophenotypic analysis of peripheral blood of these two patients confirmed involvement by mantle cell lymphoma. Three patients received chemotherapy before referral to this institution. Four of five patients received chemotherapy at our hospital. Two (Cases 1 and 2) patients died 22 and 60 months after initial diagnosis, respectively. Three patients remain alive with 3, 10, and 36 months of clinical follow-up.
Morphologic and Immunophenotypic Findings
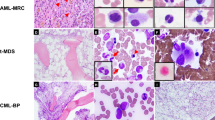
All five patients had bone marrow involvement by mantle cell lymphoma. Involvement ranged from 40 to 100% of the total cellularity, with an interstitial and diffuse pattern. A minor nodular pattern was observed in two cases. The neoplastic cells in bone marrow aspirate smears had blastoid features: three pleomorphic and two classic (Table 2). The cells ranged from medium to large in size. Nuclear contours varied from slightly to markedly irregular. The neoplastic cells had immature nuclear chromatin, and nucleoli were identified, ranging from small to prominent. One case with immature chromatin and prominent central nucleoli is shown in Figure 1. Mitotic figures were easily identified in all cases.
Peripheral blood smears, available for review in three cases, showed neoplastic cells with morphologic features similar to those observed in corresponding bone marrow aspirate smears. One case with immature chromatin and irregular nuclear contours is shown in Figure 2.
Lymph node biopsy specimens from three patients were reviewed. The pattern of the neoplasm was diffuse in two cases and nodular in one case. In one neoplasm, a starry-sky pattern was present, as shown in Figure 3. The neoplastic cells were larger with more open chromatin, as compared with typical cases of mantle cell lymphoma, and mitotic figures were numerous (Fig. 3B). One patient underwent endoscopy of the upper and lower gastrointestinal tract, and mantle cell lymphoma was histologically shown to involve the colon.
Flow-cytometric immunophenotypic analysis in all cases showed that the neoplastic cells were positive for monotypic surface immunoglobulin light chain (kappa 3, lambda 2), CD5, CD19, CD20, and FMC7. The neoplastic cells were negative for CD3, CD4, CD7, CD10, and CD23.
Immunohistochemical studies showed overexpression of cyclin D1 with a nuclear pattern in all five cases. Ki-67 and CD43 were also assessed in all cases. We semiquantified the number of Ki-67–positive cells into four groups: <25%, 25% to <50%, 50% to <75%, and ≥75%. Three cases had >75%, one had 50% to <75%, and one had 25% to <50% positive cells. Three of five tumors were positive for CD43. Other antibodies were assessed in a nonsystematic manner. The neoplastic cells were also positive for CD5 (n = 3), CD20 (n = 2), and BCL-2 (n = 1) and were negative for CD3 (n = 3), CD10 (n = 1), CD23 (n = 1), CD30 (n = 1), and TdT (n = 1).
Cytogenetic Findings
The cytogenetic findings are listed in Table 2. Complex karyotypes with multiple abnormalities were present in all five cases. All cases had cytogenetic evidence of the t(11;14)(q13;q32) and abnormalities involving 8q24, including one with the t(8;14)(q24;q32), one with the t(2;8)(q13;q24), and three with add(8)(q24).
Fluorescence In Situ Hybridization Findings
Fluorescence in situ hybridization analysis was performed on all cases, and the results are shown in Table 3. In Case 5, IgH/myc fusion signals were detected in 95 of 100 nuclei analyzed, confirming the presence of the t(8;14) involving c-myc (Fig. 4A). In Case 4, 50% of the nuclei analyzed had split c-myc signals consistent with the t(2;8) disrupting the c-myc gene (Fig. 4B). In the three cases with add(8)(q24), in Case 1, 60% of nuclei analyzed had split c-myc signals consistent with a translocation disrupting the c-myc locus. The partner chromosome in this case is not chromosome 14 but is otherwise unknown. In the other two cases, 44% (Case 2) and 76% (Case 3) of nuclei analyzed had three copies of c-myc.
Results of fluorescence in situ hybridization. A, Case 5: IgH/c-myc fusion signals are identified. (LSI IgH/MYC/CEP8 probe; c-myc is red, IgH is green, and CEP8 is blue, 1000 ×). B, Case 4: split c-myc signals are identified (LSI MYC Dual Color Breakapart Probe; intact c-myc gene is yellow, 1000 ×).
DISCUSSION
We describe five cases of mantle cell lymphoma in which conventional cytogenetic analysis revealed complex karyotypes, including the t(11;14)(q13;q32) and abnormalities of 8q24: one t(8;14)(q24;q32), one t(2;8)(p13;q24), and three add(8)(q24). We performed FISH analysis on all cases and showed involvement of the c-myc gene. C-myc was fused with IgH in the case with the t(8;14) and split apart in two cases, including the case with the t(2;8), consistent with a translocation disrupting the c-myc locus. Three copies of c-myc were observed in two other cases. This is the largest group of mantle cell lymphoma cases with 8q24 abnormalities reported in the literature, and this is the second study to prove involvement of the c-myc gene.
Ten cases of mantle cell lymphoma with the t(11;14) and abnormalities of 8q24 have been reported previously and are listed in Table 4 (14, 15, 16, 17, 18, 19). In most cases, the diagnosis of mantle cell lymphoma was established using the revised European-American lymphoma (REAL) or World Health Organization (WHO) classifications (1). Two cases were designated originally as intermediate lymphocytic lymphoma and small lymphocytic lymphoma. In retrospect, these cases are also likely to be mantle cell lymphoma as these tumors were designated as intermediate lymphocytic lymphoma or, less commonly, as small lymphocytic lymphoma, in older classifications (23). One additional case was designated originally as diffuse histocytic lymphoma using the Rappaport scheme (14). We suspect that this case was a high-grade variant of mantle cell lymphoma because some blastoid variants can have large, pleomorphic cells, as is recognized in the WHO classification (1). In all 10 cases, the cytogenetic abnormalities were complex. The 8q24 abnormalities included two t(8;14)(q24;q32), one t(2;8)(p12;q24), one t(2;8)(p11;q24), one +der(8) t(8;?)(q24;?), one t(8;9)(q24;p13), one dic(8;9)(q24;p24), one add(8)(q24), one dup(8)(q24q13), and one ins(8;14)(q24;q24q32). In the largest of these studies, Au and colleagues (18) used Southern blot methods and a c-myc probe in two of four cases but were unable to demonstrate involvement of c-myc. Recently, Vaishampayan and coworkers (19) have proven, by FISH, the presence of c-myc involvement in two cases. In one case with the t(8;14), IgH/myc fusion signals were identified. In a second case with dup(8)(q24q13), an extra copy of the c-myc gene was observed.
Au and colleagues (18) correlated 8q24 abnormalities with blastoid morphology and an aggressive clinical course in three of four cases of mantle cell lymphoma. Similarly, one case reported by Vaishampayan et al. (19) was blastoid, and the neoplasm classified as diffuse histocytic lymphoma in the study by Kaneko et al. (14), in retrospect, is also probably a pleomorphic blastoid variant of mantle cell lymphoma. In our study, all five cases had blastoid morphology: two classic and three pleomorphic type, as defined in the WHO classification (1). Thus, 8q24 abnormalities appear to correlate with blastoid morphologic variants of mantle cell lymphoma. A study by Hernandez and colleagues (24) also may be relevant. They showed higher levels of c-myc mRNA in mantle cell lymphoma cases with blastoid morphology.
Of interest, four of five patients in this study had leukemic involvement, and three patients had marked lymphocytosis, with counts ranging from 40.9 to 392.9 × 109/L. A fourth patient presented initially with a total white blood cell count of 21 × 109/L that subsequently rose to 200 × 109/L. These observations suggest that 8q24 abnormalities also may be involved in the pathogenesis of leukemic involvement. In a recent study by Martinez-Climent and colleagues (25), 8p deletion was correlated with leukemic involvement. Although 8p is relatively distant from 8q24, genetic abnormalities in either cis or trans configuration can act over long distances. Thus, it is possible that the 8q24 abnormalities in these five mantle cell lymphomas and the 8p deletions in the mantle cell lymphomas reported by Martinez-Climent and colleagues (25) may share similar mechanisms involved in the pathogenesis of marked leukemic involvement.
Although the t(11;14) is thought to be important in the pathogenesis of mantle cell lymphoma, it appears not to be sufficient for lymphomagenesis alone. Studies of transgenic mice with overexpression of cyclin D1 have failed to develop mantle cell lymphoma (4, 5). In addition, cytogenetic studies of mantle cell lymphomas have rarely shown the t(11;14) as a sole abnormality in these neoplasms (6, 7, 8, 9, 10, 11, 12, 13). In our experience, >95% of mantle cell lymphomas have additional karyotypic abnormalities (13), and all five cases in this study had complex karyotypes. Thus, it is likely that cyclin D1 overexpression combined with other abnormalities played a role in the pathogenesis of these cases. Transgenic mice models support this view, showing that double-transgenic mice with coexpression of cyclin D1 and either n-myc or l-myc demonstrate rapid development of lymphomas, suggesting that these genes cooperate in lymphomagenesis (5). Whether the c-myc abnormalities in the five cases of mantle cell lymphoma that we report were involved in pathogenesis or disease progression is unknown.
The role of c-myc in blastoid transformation of mantle cell lymphoma is unknown but presumably is similar to its role in Burkitt’s lymphoma, at least in those cases with the t(8;14) and t(2;8). The t(8;14) occurs in approximately 85%, and the t(2;8), one of the variant translocations, is found in an additional 5–10% of cases of Burkitt lymphoma. As a consequence of these translocations, the c-myc gene comes under the influence of enhancers of the immunoglobin loci, resulting in c-myc overexpression. The c-myc gene encodes a transcription factor that binds to DNA in a sequence-specific fashion and is involved in many cellular processes, including cell cycle progression, differentiation, and apoptosis (26, 27). The oncogenicity of c-myc in Burkitt’s lymphoma and in mantle cell lymphomas with the t(8;14) or t(2;8) is most likely related to its ability to stimulate cell proliferation and increase cellular metabolism. The role of the add(8)(q24) in the other three cases of mantle cell lymphoma in this study is unknown. In one case with split c-myc signals shown by FISH, most likely this case had a translocation involving c-myc in which the partner chromosome was not morphologically recognizable by conventional cytogenetics. In the other two cases, each with three c-myc copies consistent with c-myc duplication, the role of c-myc is unknown.
In summary, we describe five cases of blastoid mantle cell lymphoma with the t(11;14) and abnormalities of the 8q24 locus and have proven c-myc involvement in all cases. Only 10 cases of mantle cell lymphoma with the t(11;14) and 8q24 abnormalities have been reported previously, including 2 with the t(2;8) and 2 with the t(8;14). Collaboration between cyclin D1 and c-myc may have played a role in either lymphomagenesis or morphologic transformation in these neoplasms. Abnormalities of c-myc may also correlate with marked leukemic involvement.
References
Swerdlow SH, Nathwani BN, Berger F, Piris MA, Isaacson PI, Harris NL, et al. Mantle cell lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. World Health Organization classification of tumours. Lyon, France: IARC Press; 2001. p. 168–170.
Lai R, Medeiros LJ . Pathologic diagnosis of mantle cell lymphoma. Clin Lymphoma 2000; 1: 197–206.
Swerdlow SH, Williams ME . From centrocytic to mantle cell lymphoma: a clinicopathologic and molecular review of 3 decades. Hum Pathol 2002; 33: 7–20.
Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM . Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J 1994; 13: 2124–2130.
Lovec H, Grzeschiczek A, Kowalski M, Moroy T . Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J 1994; 13: 3487–3495.
Bigoni R, Cuneo A, Milani R, Roberti MG, Bardi A, Rigolin GM, et al. Secondary chromosome changes in mantle cell lymphoma: cytogenetic and fluorescence in situ hybridization studies. Leuk Lymphoma 2001; 40: 581–590.
Bentz M, Plesch A, Bullinger L, Stilgenbauer S, Ott G, Muller-Hermelink HK, et al. t(11;14)-positive mantle cell lymphomas exhibit complex karyotypes and share similarities with B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer 2000; 27: 285–294.
Cuneo A, Bigoni R, Rigolin GM, Roberti MG, Bardi A, Piva N, et al. Cytogenetic profile of lymphoma of follicle mantle lineage: correlation with clinicobiologic features. Blood 1999; 93: 1372–1380.
Espinet B, Sole F, Woessner S, Bosch F, Florensa L, Campo E, et al. Translocation (11;14)(q13;q32) and preferential involvement of chromosomes 1, 2, 9, 13, and 17 in mantle cell lymphoma. Cancer Genet Cytogenet 1999; 111: 92–98.
Johansson B, Mertens F, Mitelman F . Cytogenetic evolution patterns in non-Hodgkin’s lymphoma. Blood 1995; 86: 3905–3914.
Khokhar MT, Brito-Bapapulle V, Matutes E, Catovsky D . Cytogenetic abnormalities in the leukemic phase of non-Hodgkin’s lymphoma. Cancer Genet Cytogenet 1995; 83: 18–24.
Wlodarska I, Pittaluga S, Hagemeijer A, De Wolf-Peeters C, Van den Berghe H . Secondary chromosome changes in mantle cell lymphoma. Haematologica 1999; 84: 594–599.
Onciu M, Schlette E, Medeiros LJ, Abruzzo LV, Keating M, Lai R . Cytogenetic findings in mantle cell lymphoma. Cases with a high level of peripheral blood involvement have a distinct pattern of abnormalities. Am J Clin Pathol 2001; 116: 886–892.
Kaneko Y, Rowley JD, Variakojis D, Haren JM, Ueshima Y, Daly K, et al. Prognostic implications of karyotype and morphology in patients with non-Hodgkin’s lymphoma. Int J Cancer 1983; 32: 683–692.
Bloomfield CD, Arthur DC, Frizzera G, Levine EG, Peterson BA, Gajl-Peczalska KJ . Nonrandom chromosome abnormalities in lymphoma. Cancer Res 1983; 43: 2975–2984.
Leroux D, Le Marc’Hadour F, Gressin R, Jacob MC, Keddari E, Monteil M, et al. Non-Hodgkin’s lymphoma with t(11;14)(q13;q32): a subset of mantle zone/intermediate lymphocytic lymphoma? Br J Haematol 1991; 77: 346–353.
Tirier C, Zhang Y, Plendl H, Weber-Matthiesen K, Langer W, Heit W, et al. Simultaneous presence of t(11;14) and a variant Burkitt’s translocation in the terminal phase of a mantle cell lymphoma. Leukemia 1996; 10: 346–350.
Au W, Horsman D, Viswanatha DS, Connors JM, Klasa RJ, Gascoyne RD . 8q24 translocations in blastic transformation of mantle cell lymphoma. Haematologica 2000; 85: 1225–1227.
Vaishampayan UV, Mohamed AN, Dugan MC, Bloom RE, Palutke M . Blastic mantle cell lymphoma associated with Burkitt’s-type translocation and hyperploidy. Br J Haematol 2001; 115: 66–68.
Schlette E, Bueso-Ramos C, Giles F, Glassman A, Hayes K, Medeiros LJ . Mature B-cell leukemias with more than 55% prolymphocytes. A heterogeneous group that includes an unusual variant of mantle cell lymphoma. Am J Clin Pathol 2001; 115: 571–581.
Borowitz MJ, Guenther KL, Shults KE, Stelzer GT . Immunophenotyping of acute leukemia by flow cytometric analysis: use of CD 45 and right-angle light scatter to gate on leukemia blasts in three-color analysis. Am J Clin Pathol 1993; 100: 534–540.
Mitelman F, editor. ISCN 1995: an international system for human cytogenetic nomenclature 1995. Basel, Switzerland: Karger, 1995.
Medeiros LJ, Van Krieken J, Jaffe ES, Raffeld M . Association of bcl-1 rearrangements with lymphocytic lymphoma of intermediate differentiation. Blood 1990; 76: 2086–2090.
Hernandez L, Hernandez S, Bea S, Pinyol M, Ferrer A, Bosch F, et al. c-myc mRNA expression and genomic alteration in mantle cell lymphomas and other nodal non-Hodgkin’s lymphomas. Leukemia 1999; 13: 2087–2093.
Martinez-Climent JA, Vizcarra E, Sanchez D, Blesa D, Marugan I, Benet I, et al. Loss of a novel tumor suppressor gene locus at chromosome 8p is associated with leukemic mantle cell lymphoma. Blood 2001; 98: 3479–3482.
Boxer LM, Dang CV . Translocations involving c-myc and c-myc function. Oncogene 2001; 20: 5595–5610.
Hecht JL, Aster JC . Molecular biology of Burkitt’s lymphoma. J Clin Oncol 2000; 18: 3707–3721.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hao, S., Sanger, W., Onciu, M. et al. Mantle Cell Lymphoma with 8q24 Chromosomal Abnormalities: a Report of 5 Cases with Blastoid Features. Mod Pathol 15, 1266–1272 (2002). https://doi.org/10.1097/01.MP.0000037310.82136.99
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.MP.0000037310.82136.99
Keywords
This article is cited by
-
Smoldering mantle cell lymphoma
Journal of Experimental & Clinical Cancer Research (2017)
-
Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms
Modern Pathology (2017)
-
Increased MYC copy number is an independent prognostic factor in patients with diffuse large B-cell lymphoma
Modern Pathology (2017)
-
The clinical significance of 8q24/MYC rearrangement in chronic lymphocytic leukemia
Modern Pathology (2016)
-
Diagnostic and predictive biomarkers for lymphoma diagnosis and treatment in the era of precision medicine
Modern Pathology (2016)