Abstract
Despite low sensitivity (around 60%), cytomorphologic examination of urine specimens represents the standard procedure in the diagnosis and follow-up of bladder cancer. Although color is information-rich, morphologic diagnoses are rendered almost exclusively on the basis of spatial information. We hypothesized that quantitative assessment of color (more precisely, of spectral properties) using liquid crystal-based spectral fractionation, combined with genetic algorithm-based spatial analysis, can improve the accuracy of traditional cytologic examination. Images of various cytological specimens were collected every 10?nm from 400 to 700?nm to create an image stack. The resulting data sets were analyzed using the Los Alamos-developed GENetic Imagery Exploitation (GENIE) package, a hybrid genetic algorithm that segments (classifies) images using automatically ‘learned’ spatio-spectral features. In an evolutionary fashion, GENIE generates a series of algorithms or ‘chromosomes’, keeping the one with best fitness with respect to a user-defined training set. First, we tested the system to determine if it could recognize malignant cells using artificial cytology specimens constructed to completely avoid the requirement for human interpretation. GENIE was able to differentiate malignant from benign cells and to estimate their relative proportions in controlled mixtures. We then tested the system on routine cytology specimens. When targeted to detect malignant urothelial cells in cytology specimens, GENIE showed a combined sensitivity and specificity of 85 and 95%, in samples drawn from two separate institutions over a span of 4 years. When trained on cases initially diagnosed as ‘atypical’ but with unequivocal follow-up by biopsy, surgical specimen or cytology, GENIE showed efficiency superior to the cytopathologist with respect to predicting the follow-up result in a cohort of 85 cases. We believe that, in future, this type of methodology could be used as an ancillary test in cytopathology, in a manner analogous to immunostaining, in those situations when a definitive diagnosis cannot be rendered based solely on the morphology.
Similar content being viewed by others
Main
Urothelial carcinoma is diagnosed and followed by cytological evaluation of voided urine and bladder washings.1, 2 The sensitivity of these tests is moderate, ranging between 60 and 80%.3, 4 Low-grade tumors offer an even greater challenge to the cytopathologist,5 resulting in the use of such terminology as ‘atypical urothelial cells’, leading to suboptimal patient management.6
Morphologic diagnoses in cytopathology are primarily driven by spatial relationships, while color-based information is relegated to a very minor role. This may be because the human eye has limited spectral resolution, viewing the world in broad, overlapping spectral bands of red, green and blue. If the spectral properties of cytological specimens are important, this fact can only be assessed using devices capable of imaging with spectral precision. This excludes conventional red–green–blue (RGB) cameras, which have no more spectral resolving power than the human visual system they are designed to emulate. Spectral imaging, on the other hand, is a relatively novel technique capable of quantitatively measuring optical spectra on a pixel-by-pixel basis, and therefore capturing differences normally overlooked by the human eye.7 Previous studies have shown that spectral analysis has high information content, albeit less than spatial information.8 Thus, combination of spectral imaging with spatial analysis (morphology) could maximize information obtainable from routine pathology slides and could serve as a valuable ancillary test in diagnostic pathology.
GENetic Imagery Exploitation (GENIE) is a recently developed artificial intelligence platform that allows optimization of image analysis algorithms through selection of learned spatio-spectral features.9 This system was originally conceived at the Los Alamos National Laboratory for the analysis of satellite images of the surface of the earth.10 GENIE is a hybrid learning system that combines a genetic algorithm that searches in a space of image-processing operations for a set that can produce suitable feature planes with a more conventional classifier that uses those feature planes to output a final classification. In an evolutionary fashion, GENIE generates a series of mathematical algorithms named ‘chromosomes’ by John Holland, the inventor of this system of analysis. GENIE then assigns an individual fitness score related to how well each chromosome classifies a set of training images. Each chromosome is composed of a variable number of mathematical operators that were named ‘genes’, in analogy to the biological system on which the mathematical model was based. The genes are then interpreted serially to result in a chromosome's fitness score. At the end of each generation, GENIE selects chromosomes to participate in processes of crossover and mutation, with the probability of selection being based on their fitness. This cycle is repeated many times until optimal fitness or convergence occurs. When compared to other supervised classifiers for multispectral image feature analysis, GENIE outperformed them all in almost every task tested, indicating higher sensitivity and generalization abilities.11
Here, we first applied the GENIE hybrid genetic algorithm to multispectral images obtained from artificially produced Papanicolaou-stained cytology slides containing defined mixtures of benign and malignant colonic cells, to test if the system was able to distinguish these classes, free of human intervention. Then, we tested GENIE in the analysis of routine urine cytology slides, which were prepared at different time-periods and at separate institutions. Finally, we evaluated GENIE's performance on urine cytology specimens that were initially called ‘atypical’ by the cytopathologist, to evaluate its potential use as an ancillary test.
Materials and methods
Preparation of Cytology Specimen of Defined Composition
Suspensions of benign and malignant colonic epithelial cells were obtained by manually scraping the mucosal surface of a fresh colon adenocarcinoma resection specimen received at the Department of Pathology at Yale. The scraped material was immediately suspended in RPMI medium and gently dispersed using a glass Dounce homogenizer. An aliquot of the suspension was used to determine cell count using a hemocytometer. Viability was at least 90% as determined by trypan blue dye exclusion.12 Aliquots from each suspension were mixed in 11 ThinPrep™ vials containing 20?ml of PreservCyt® preservative at different final malignant/benign cell ratios (from 0/100 to 100/0 in 10% increments), and with a total cell concentration of 1 × 105?cells/ml. A ThinPrep™ slide was prepared from each vial in a 2000 ThinPrep™ processor (Cytyc, Boxborough, MA, USA). All slides were stained together with Papanicolaou stain. In all, 20 random, high-power (× 400) fields containing cells or cell clusters were selected for multispectral image acquisition (see the section; Data Collection). Five extra additional areas were selected in the slides containing 100 and 0% tumor cells, which were used for GENIE training purposes.
Urine Cytology Specimen Selection
Papanicolaou-stained urine cytology ThinPrep™ slides were collected from archival material at the Departments of Pathology of Yale-New Haven Hospital and University of Massachusetts Medical School. Representative areas containing benign, atypical or malignant urothelial cells were selected by a cytopathologist for image data acquisition. The collection of this material and associated clinical information was approved by the Yale Human Investigation Committee in protocol #8219.
Data Collection
High-power field (× 400) multispectral images were acquired from selected areas of the ThinPrep™ cytology slides at 420–700?nm (10-nm intervals) using a BH-2 Olympus light microscope (Olympus America, Melville, NY, USA) equipped with a CRI VariSpec™ and VIS2-CM liquid crystal tunable filter (CRI, Woburn, MA, USA). The tunable filter was coupled to a Retiga 1300 monochrome CCD camera (Quantitative Imaging, Burnaby, BC, Canda). Both CCD camera and VariSpec were controlled by CRI acquisition software (CRI, Woburn, MA, USA). Exposure time for each slide and wavelength was calculated for each slide prior to each acquisition. Background information was subtracted by acquiring an area with no cells (one per slide) and flat-fielding over the area of interest. Digital images were produced as 29-stack arrays of 896 × 768 pixels at 8-bit resolution (metafiles).
GENIE Training and ‘Chromosome’ Generation
Training multispectral image data were introduced into the GENIE analysis by means of the ALADDIN Java-based graphic tool. ALADDIN allows the analyst to select those areas in the imported training images to be considered as ‘feature’ (eg malignant cells) and ‘nonfeature’ (eg benign cells) by manually painting those areas as a colored overlay (green and red, respectively). These transformed images (GENIE training files) are then imported in the GENIE training dialog box window. Before starting a training session, the learning system parameters were specified in terms of number of chromosomes per generation (60), maximum number of genes in each algorithm (20), backend discrimination (Fisher), crossover mechanism (Singlepoint), crossover rate (0.9), mutation parameter rate (0.25), gene mutation rate (0.6), fitness metric (Hamming), thresholding (Intelligent), selection rate (Tournament 3), elite fraction (0.1) and end-point fitness (1000). The allowed number of generations was different for each training session, ranging between 300 and 500.
Validation of GENIE ‘Chromosomes’
Multispectral image data from the test sets were subjected to mathematical transformation by means of those algorithms (chromosomes) previously generated in the training process. The modified result images could be viewed as green (feature) and red (nonfeature) areas overlaying the original images (used for orientation purposes), or as white–black binary images (used for scoring purposes). Note that the choice of the terms ‘gene’ and ‘chromosome’ is that of the designers of genetic algorithms. Thus, although they may be confusing in a biological setting, we did not attempt to change these terms since they are the standard terminology for genetic algorithms. Since the image analysis of microscopic slides by the GENIE's chromosomes is not carried out in the context of cells, tissues or disease, but looking at pixel spectral quality and their spatial mathematical integration, we needed, at least initially, to design a way to translate or ‘score’ its results back to the clinical context. We chose to do it by calculating the GENIE index value, which represents the proportion of pixels inside the selected object of study (nuclei, whole cell, cell cluster, etc.) that is recognized as ‘feature’. Based on these data, receiver operator characteristic (ROC) curves were constructed on Excel Analyze-it® software (Leeds, UK). Combined sensitivity and specificity were calculated using increasing levels of GENIE index as cutoff values.
Results
Detection of Malignant Cells in Defined-Composition Cytology Specimens
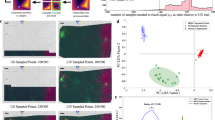
This first experiment was performed to determine if GENIE could recognize cancer cells from normal cells in regular cytology preparations, however free of subjective pathologist interpretation. For this purpose, we created artificial cytology specimens containing benign and malignant colonic epithelial cells mixed at known different ratios. In this particular case, we chose to use colon and not bladder mostly for practical reasons and to avoid possible interference due to inadvertent field effect, which is a common feature of urothelial neoplasms. The purpose of this experiment was to test if GENIE could differentiate two distinct populations of cells (benign and malignant) from routinely processed ThinPrep® cytology preparations. From each mixture, one ThinPrep™ slide was prepared, and all slides were stained together using the Papanicolaou stain. GENIE training was performed with five high-power (× 400) multispectral images (420–700?nm, 10-nm intervals) acquired from the slides containing 0 and 100% tumor cells (10 multispectral images in total). The training images were created by manually marking every cell or cell cluster from the images containing 0% tumor cells as ‘nonfeature’ or benign (by overlaying red color), and every one from the images containing 100% tumor cells as ‘feature’ or malignant (by overlaying green color). These GENIE training files were imported into the training session dialog box and run for 300 generations. This process generated a chromosome termed #004345, showing fitness with respect to the training files set of 835 (a score of 1000 would be consistent with the chromosome classifying every pixel in the training data set correctly). The chromosome was composed of three operators aligned in the following order: [ASF_CLOP rD5 wS1 2 1][SUBP rD29 rD24 wS2][SQRT rD12 wS0]. ASF_CLOP is an alternating sequential filter (close–open), which basically performs an open–close operation on data plane D5 (470?nm) at increasing sizes using a circular structuring element, writing the result in scratch plane S1. SUBP is a subtract plane operator that subtracts data plane D24 (660?nm) from data plane D29 (700?nm). SQRT performs pixel-by-pixel square-root calculation in data plane D12 (540?nm) and writes the result in scratch S0. The results from each scratch plane are added into a feature plane and a threshold value is calculated. In order to test this chromosome, new multispectral test images were acquired from the slides containing different proportions of malignant to benign colonic epithelial cells (20 areas per slide, including new images from the slides containing 100 and 0% malignant cells), and run in GENIE using chromosome #004345. Visual comparison of the GENIE result testing images from the slides containing benign or malignant cells only showed that most of the pixels corresponding to tumor cells were clearly identified as ‘feature’ (green color overlaying the original images), while those corresponding to benign colonic epithelial cells were ignored (red color, see Figure 1a). The same visual analysis is not possible in those cases where mixtures of both malignant and benign colonic epithelial cells are present, due to the fact that they are hard to recognize on the original Papanicolaou stained images. However, since the actual proportion of cells in each slide is known, we could evaluate GENIE's performance by plotting the proportion of total cell-associated pixels per image that are identified as ‘feature’. As the relative number of malignant cells in the original mixture increases, so would the percentage of the total cellular pixels (GENIE index) in the digital images. This assumes that the cell mixtures are homogeneously layered on the slides by the ThinPrep processor, which is not the exact case, and this may be our largest source of error. However, these inequalities should be partially compensated by increasing the number of images taken per slide (we used 20 images per slide). Also, since the surface occupied by cells is variable from image to image, we could not just use the total number of pixels identified in each image, but should relate it to the total cell surface (in pixels), excluding the background. In order to do this in an objective fashion (again free of pathologist intervention), we created chromosome #background_092603_000004 that identifies background and ignores cells. Chromosome #background_092603_000004 was generated using the same training images as chromosome #004345, but selecting areas of background as ‘feature’ and areas containing cells (either benign or malignant) as ‘nonfeature’. Chromosome #background_092603_000004 had the following sequence: [RANGE rD24 wS2 2 1][MIN rS2 rD6 wS0][ADDP rD19 rD8 wS1][ASF_OPCL rD22 wS2 2 1]. Based on this sequence, the algorithm performs a combination of pixel-neighborhood operations (RANGE; ASF_OPCL), addition between two data planes (ADDP) and a logical operation (MIN). We then combined the results of both chromosomes in each validation image, so as to mask the background, and exclusively score the results on cell-associated pixels. When we plotted the average GENIE index, using 20 random areas per slide (which represents approximately 5% of the operational surface in the ThinPrep™ slide), we observed a fairly linear relation between the percentage of tumor cells known to be in the original suspensions and the mean GENIE index (correlation coefficient: 0.84) (Figure 1b).
Detection of malignant cells in cytology specimens of defined composition using GENIE chromosome #004345. GENIE results are displayed on the right as binary green (feature) and red (nonfeature) images, superimposed to the original image (which is better appreciated on the left). (a) (A–D) Testing images taken from the slide containing no colon adenocarcinoma cells. (a) (E–H) Testing images from the 100% colon adenocarcinoma slide. (b) Quantification of malignant colonic epithelial cells in mixtures of defined composition. Every data point is the average of 20 high-power fields (× 400) from each slide, which was prepared from vials containing a defined proportion of malignant cells.
Detection of Malignant Urothelial Cells in Routine Urine Cytology Specimens
Our next step was to create a chromosome able to detect malignant urothelial cells in routine urine specimens. We trained GENIE with 12 Papanicolaou stained ThinPrep™ urine cytology slides received in the Department of Pathology at the Yale-New Haven Hospital between 1996 and 1997. From these slides, 38 high-power multispectral images were acquired and imported into the ALADDIN platform. In total, 161 nuclei from benign urothelial cells and 70 nuclei from malignant urothelial cells were marked by a cytopathologist as ‘nonfeature’ and ‘feature’, respectively. Cells showing atypia, but lacking the definitive features of malignancy, as well as inflammatory cells and degenerated urothelial cells, were not selected. GENIE training (500 generations) using these files produced a single best chromosome, #025867, containing four genes, which was evaluated on two validation sets. The first set was constructed from 17 unique patients’ slides with a diagnosis of urothelial carcinoma received in the Department of Pathology at Yale-New Haven Hospital between 1998 and 1999. The set contained a total of 190 benign urothelial cells and 178 malignant urothelial cells. GENIE identified malignant cells by overlaying green color over their nuclear areas (Figure 2, center column). Using 90% of the nuclear area being identified as ‘feature’ as the cutoff value (GENIE index value: 0.9), this chromosome classified malignant and benign urothelial cells with a sensitivity and specificity of 87 and 96%. Atypical or degenerate cells were not used in the calculation, since they were excluded from the training set. Inflammatory cells were also excluded; however, this chromosome did not identify neutrophils as ‘feature’. The second validation set was composed of eight cases (six positive for urothelial carcinoma and two negative for malignancy) from the Department of Pathology at University of Massachusetts Medical School. The Papanicolaou staining protocol routinely used at both institutions is similar. This test set comprised 121 benign urothelial cells and 40 malignant urothelial cells. Against this set, chromosome #025867 demonstrated a sensitivity and specificity of 85 and 96%.
Detection of malignant urothelial cells in routine urine cytology slides using GENIE. Original images are displayed on the left side of the figure. The center column corresponds to GENIE result images using chromosome #025867. This chromosome identifies the nuclei of urothelial carcinoma cells by overlaying green color over its surface. The right column shows the performance of chromosome #025867 when spectral data planes D13 and D15 are switched (in an analogous fashion to transposition mutation).
Analysis of the sequence of chromosome #025687 showed that it is composed of four genes: [ASF_OPCL rD15 wS2 6 1][NEG_TH rS2 wS0 9 3][RANGE rD13 wS1 8 1][ASF_OPCL rS0 wS0 7 1]. In this case, only two data planes closely related in the spectra (rD15 corresponding to 570?nm and rD13 to 550?nm) were being used. This initially suggested that the analysis codified by chromosome #025687 was mainly spatial. In order to test if any spectral information was relevant for this chromosome performance, we designed an experiment in which the information from these two spectral data planes is interchanged, by manually altering their location in the metafile (image stack). As a result, the mathematical operator that originally used the 550?nm plane (RANGE) was given the 570?nm plane and vice versa (the first ASF_OPCL was performed on the 550?nm data plane). Under these new conditions (homologous to genetic transposition), chromosome #025867 misclassified several benign nuclei as malignant (Figure 2, right column), further illustrating the critical nature of the spectral component of the analysis.
Detection of Malignant Urothelial Cells in ‘Atypical’ Urine Cytology Specimens
Adjudication of cases defined as ‘atypical’ represents a critical challenge for pathologists and a potential clinical application of this technology. To investigate this possibility further, we selected 16 unique urine cytology cases received at the Yale-New Haven Hospital between 1995 and 1996 and initially diagnosed as ‘atypical’ by the cytopathologist of record. Eight slides (negative follow-up group) corresponded to cases that had at least one specimen subsequent to the index case where the cytology, biopsy or surgical specimen was diagnosed as ‘negative for malignancy’ in the following year. Cases were excluded if subsequent specimens included ‘atypical’, ‘suspicious’ or ‘positive for malignant cells’ in the diagnostic text. The other eight (positive follow-up group) had at least one specimen subsequent to the index case where the biopsy or surgical specimen was called ‘positive for malignant cells’ in the following year. A single image containing the most atypical cell cluster was used for each case to define the training set. Areas corresponding to the whole atypical clusters of the ‘follow-up positive group’ were manually designated as ‘feature’, while the ones of ‘follow-up negative group’ as ‘nonfeature’, in the ALADDIN platform. In this case, whole cells or cellular clusters were used as the object of study, instead of nuclei, due to the fact that the atypical cells of most cases in our cohort were present in the form of cell aggregates with common nuclear overlapping. By using similar training conditions as those used for chromosome #025867, GENIE generated chromosome #026897. Chromosome #026897 had the following sequence: [QTREG rD26 wS1 wS2 wS0 0.05][SQRT rD13 wS1][SADIST rD4 rD23 rD5 rD21 rD16 rD22 rD0 rD24 rS1 rD7 wS0 −1.18 −1.00 1.38 0.94 −1.21 −1.24 1.23 1.00 0.88 −0.095]. The first gene (QTREG) performs statistical analysis in relation to region size, while the second gene (SQRT) is a basic mathematical operation (square root). The third gene (SADIST) performs spectral angle calculations between neighborhood regions. The fitness of this chromosome with respect to the training set was 824, which means that 82.4% of the pixels correctly distinguished feature from nonfeature. We then prepared a completely unique validation set using images from 34 ‘negative follow-up’ and 51 ‘positive follow-up’ urine cytology cases from archival material received between 1997 and 2002. The criteria used in the selection of cases were similar to those for the training set. One atypical cell cluster per slide was selected for acquisition and each sample corresponded to a different patient. Scoring was carried out by calculation of the GENIE index in each cell cluster alone. The results are shown as a frequency distribution of GENIE index scores (Figure 3a and b). The positive follow-up group shows a significantly higher mean score than the negative follow-up. There is no cut-point that definitively separated both groups, but an ROC curve could be generated based on the number of pixels per cell cluster used to define a positive identification. The area under the ROC curve obtained with chromosome #026897 was 0.728 (Figure 4).
GENIE detection of malignancy in routine urine cytology slides previously diagnosed as ‘atypical’ by a cytopathologist. (a) Results obtained with chromosome #026897 on atypical urine cytology cases with follow-up negative. (b) Results obtained with chromosome #026897 on atypical cases with follow-up positive. (c) Results obtained with chromosome #020105 on atypical cases with follow-up negative. (d) Results obtained with chromosome #020105 on atypical cases with follow-up positive.
We attempted to improve the performance of GENIE on atypical urine cytology cases by re-training the system using a larger number of training images (32 cases, including those used in the previous training session). The assumption was that by increasing the number of training images, GENIE would generate a ‘smarter’ chromosome capable of recognizing larger number of classificatory features. Using this strategy, the resulting chromosome was #020105, which displayed the following sequence: [SANORM rD15 rD17 rD10 rD25 wS1 wS0 wS2][DILATE rD16 wS0 2 7][MIN rS1 rD2 wS1][CLIP_HI rD12 wS2 0.06][s.d. rS1 wS3 5 0][POS_TH rS2 wS1 10 3]. This is a relatively complex chromosome that performs spectral analysis (SANORM), pixel neighborhood operations (DILATE, s.d.: Standard Deviation, POS_TH: Positive Top-Hat), threshold operations (CLIP_HI: Clip-High) and a logical pixel-wise minimum (MIN). The fitness of chromosome #020105 with respect to the training set was 732, which is lower than the one corresponding to the previous chromosome (#026897) that was trained with less number of cases. However, when this chromosome was run on the test set, it performed better compared to #026897 (Figure 3c and d), with an area under the ROC curve of 0.784 (Figure 4).
Discussion
GENIE is a newly developed artificial intelligence system that performs spatio-spectral analysis of images. This is in contrast to a long history of quantitative analysis systems tested on urine specimens which all have used spatial information, and monochromatic (or panchromatic) optical density.13, 14, 15, 16 In comparison to these types of studies, GENIE increases the final information content by adding spectral data, which, based on our previous work,8 appears to contain relevant clinical information. Note that the wavelengths selected by GENIE are from a range of 29 wavelengths and are not those that would create a standard RGB image. The fact that certain wavelengths are more informative than others suggests subtleties in protein–dye interactions that are impossible to appreciate with the human eye.
Our first goal was to prove that GENIE was able to classify carcinoma cells in routine ThinPrep™ cytology slides without human adjudication or pathologist interpretation. For this purpose, we created a series of artificial cytology slides containing defined mixtures of benign and malignant colonic epithelial cells. GENIE was able to distinguish the malignant colonic cells from the benign and estimate the relative amounts present in each mixture. The results seen may underestimate the discriminatory power of GENIE since the accuracy is degraded by the inaccuracy of dilution of cells in creation of the artificial cytology specimens.
Our second step was to create a chromosome that could identify malignant urothelial cells in routine ThinPrep™ cytology specimens. For this purpose, GENIE was trained using examples of nuclei from cytologically benign and malignant urothelial cells. The chromosome obtained, termed #025867, when validated against two cohorts of cases (from different years and institutions), performed in a comparable way to the cytopathologist, showing combined sensitivities and specificities of over 85 and 95%, respectively. It is noteworthy that a critical difficulty in this work is to compare the objective results of the GENIE analysis to the subjective results of the pathologist. This was particularly true when GENIE was confronted with urine cytology cases that were classified as ‘atypical’ by the cytopathologist. Currently, there is no completely accepted gold standard that we could use to unequivocally rule out malignancy in a particular selected urothelial cluster from any given cytology cases that carry a diagnosis of ‘atypical’. An ‘atypical’ urine cytology diagnosis followed by discovery of malignancy in a cytoscopy a year later could mean either that it was not detected (by calling it atypical in the previous cytology specimen) or also that it developed after an ‘atypical, but in fact benign’ cytology specimen was obtained. One possible way to solve this issue could be the endoscopic biopsy of the urothelial mucosa at the same time with the collection of urine, or the simultaneous use of fluorescence in situ hybridization to detect chromosomal abnormalities. Prospective studies like this are just being started in our group, although they still may not offer a complete assurance, since there could be limitations in sampling. It needs to be stressed that the aim of this study is to compare this technology to the ‘real-life’ cytopathologist performance. In the present study, the use of follow-up data with cytology, biopsies and surgical specimens seemed the most reasonable approach from a logistic point of view. We limited the time frame to which the confirmatory subsequent studies had to be performed to one year from the time where the ‘atypical’ diagnosis was made. This meant that one atypical cytology case that had a follow-up malignant biopsy, for instance, 3 years later, was not included, because we do not have a practicable way to prove if in fact the tumor was in that original ‘atypical’ cytology specimen or appeared later on. An atypical cytology was called ‘follow-up negative’ only when there was a subsequent specimen called negative (also in the following year); however, if it was also followed by another atypical (in that same year period), it was rejected. In order to be consistent with our criteria of selection, if an atypical cytology specimen was followed by a negative cytology within the following year, and years later a biopsy finds tumor, it was still considered as ‘negative follow-up’, under the assumption that this last biopsy is the result of the later development of tumor. However, this last situation is present in less than 5% of the cases used for the construction of the validation set. Using these rules of classification, GENIE produced chromosome #026897, which was able to predict urothelial malignancy in a cohort of 85 patients with a combined sensitivity and specificity of approximately 67% (area under the ROC curve of 0.72).
It should also be noted that while the pathologist's report was generated looking at the whole slide, the GENIE ‘diagnosis’ was based on a single cluster per slide. Presumably, by increasing the number of cell clusters analyzed by GENIE in the validation set, the efficiency of the system should increase too. Similarly, increasing the number of clusters in the training set should also increase the accuracy of the system. We pilot-tested this idea by doubling the number of training images in a new training session. GENIE generated chromosome #020105, which showed higher sensitivity (78%) and specificity (72%) on the same specimen set with an area under the ROC curve of 0.78. Even though this number is much better that the 0.50 inferred by the diagnosis of ‘atypical’, it should be interpreted with caution when compared to the performance of a cytopathologist. In future, larger studies will be required for more accurate comparisons to pathologist performance.
Interestingly, the fitness of this new chromosome with respect to the original training set had a lower value than the one found for chromosome #026897. In addition, we noted that when chromosome #020105 was run on the validation set, the range in the GENIE index values was smaller. This suggests that even when the new chromosome was a ‘smarter’ one (since the area under the curve had improved), it was more ‘hesitant’ too (since the dynamic range of the GENIE index decreased). This indicates that, as we increase the number of training features, GENIE experiences more difficulties in finding a single algorithm that could encompass every spatio-spectral variant. This problem could possibly be solved by: (1) using different training conditions (eg increasing the number of generation cycles), (2) retraining the system with the addition of those features that were missed by the previous chromosome (in the present study, the additional cases were randomly selected) or (3) combining algorithms trained to identify more restricted features, and integrating them through a higher-order ‘cognizant’ chromosome (abstract thinking or data fusion). This last option represents an area of intensive research in modern computational analysis of remote-sensing data,17 and is expected to become the future realm of machine learning, as the search for suitable algorithms will evolve far beyond the human capabilities in a realistic time frame. We have currently begun applying this concept on our cytology samples in new studies in order to evaluate its clinical potential.
References
Messing E . Urothelial tumors of the urinary tract. In: Walsh PC, Retik AB, Vaughan ED, Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA (eds). Campbell's Urology. WB Saunders: St Louis, MO, 2002, pp 2732–2765.
Tut VM, Hildreth AJ, Kumar M, et al. Does voided urine cytology have biological significance? Br J Urol 1998;82:655–659.
Gregoire M, Fradet Y, Meyer F, et al. Diagnostic accuracy of urinary cytology, and deoxyribonucleic acid flow cytometry and cytology on bladder washings during followup for bladder tumors. J Urol 1997;157:1660–1664.
Kumar NU, Dey P, Mondal AK, et al. DNA flow cytometry and bladder irrigation cytology in detection of bladder carcinoma. Diagn Cytopathol 2001;24:153–156.
Curry JL, Wojcik EM . The effects of the current World Health Organization/International Society of Urologic Pathologists bladder neoplasm classification system on urine cytology results. Cancer 2002;96:140–145.
Renshaw AA . Subclassifying atypical urinary cytology specimens. Cancer 2000;90:222–229.
Farkas DL, Du C, Fisher GW, et al. Non-invasive image acquisition and advanced processing in optical bioimaging. Comput Med Imaging Graph 1998;22:89–102.
Jaganath R, Angeletti C, Levenson R, et al. Diagnostic classification of urothelial cells in urine cytology specimens using exclusively spectral information. Cancer 2004;102:186–191.
Perkins SJ, Theiler JP, Brumby SP, et al. GENIE: A hybrid genetic algorithm for feature classification in multi-spectral images. Proc SPIE 2000;4120:52–62.
Brumby SP, Theiler JP, Perkins SJ, et al. Investigation of image feature extraction by a genetic algorithm. Proc SPIE 1999;3812:24–31.
Harvey NR, Theiler JP, Brumby SP, et al. Comparison of GENIE and conventional supervised classifiers for multispectral image feature extraction. IEEE Trans Geosci Remote Sensing 2002;40:393–404.
Jauregui HO, Hayner NT, Driscoll JL, et al. Trypan blue dye uptake and lactate dehydrogenase in adult rat hepatocytes—freshly isolated cells, cell suspensions, and primary monolayer cultures. In Vitro 1981;17:1100–1110.
Koss LG, Bartels PH, Wied GL . Computer-based diagnostic analysis of cells in the urinary sediment. J Urol 1980;123:846–849.
Sherman AB, Koss LG, Wyschogrod D, et al. Bladder cancer diagnosis by computer image analysis of cells in the sediment of voided urine using a video scanning system. Anal Quant Cytol Histol 1986;8:177–186.
Van der Poel HG, Van Balken MR, Schamhart DH, et al. Bladder wash cytology, quantitative cytology, and the qualitative BTA test in patients with superficial bladder cancer. Urology 1998;51:44–50.
de la Roza GL, Hopkovitz A, Caraway NP, et al. DNA image analysis of urinary cytology: prediction of recurrent transitional cell carcinoma. Mod Pathol 1996;9:571–578.
Harvey NR, Perkins SJ, Pope PA, et al. Investigation of automated feature extraction using multiple data sources. Proc SPIE 2003;5099:190–200.
Acknowledgements
This work is supported by grants from the Patrick and Catherine Weldon Donaghue Foundation and US Army Breast Cancer Research Program Idea Award DAMD17-02-1-0634 to David Rimm and by the NIH R44 CA88684 Phase II Supplement to Richard Levenson at CRI, Inc., and by the American Society of Cytopathology Research Seed Award to Cesar Angeletti.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Angeletti, C., Harvey, N., Khomitch, V. et al. Detection of malignancy in cytology specimens using spectral–spatial analysis. Lab Invest 85, 1555–1564 (2005). https://doi.org/10.1038/labinvest.3700357
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700357