Abstract
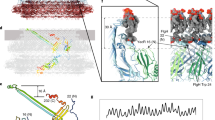
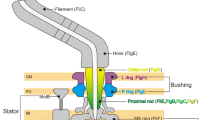
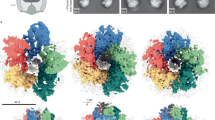
Many motile species of bacteria are propelled by flagella, which are rigid helical filaments turned by rotary motors in the cell membrane1,2,3. The motors are powered by the transmembrane gradient of protons or sodium ions. Although bacterial flagella contain many proteins, only three—MotA, MotB and FliG—participate closely in torque generation. MotA and MotB are ion-conducting membrane proteins that form the stator of the motor. FliG is a component of the rotor, present in about 25 copies per flagellum. It is composed of an amino-terminal domain that functions in flagellar assembly and a carboxy-terminal domain (FliG-C) that functions specifically in motor rotation. Here we report the crystal structure of FliG-C from the hyperthermophilic eubacterium Thermotoga maritima. Charged residues that are important for function, and which interact with the stator protein MotA4,5, cluster along a prominent ridge on FliG-C. On the basis of the disposition of these residues, we present a hypothesis for the orientation of FliG-C domains in the flagellar motor, and propose a structural model for the part of the rotor that interacts with the stator.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Blair, D. F. How bacteria sense and swim. Annu. Rev. Microbiol. 49, 489–522 (1995).
Macnab, R. M. in Escherichia coli and Salmonella typhimurium: Cellular nd Molecular Biology (eds Neidhardt, F. C.) 123–146 (American Society for Microbiology, Washington DC, (1996).
DeRosier, D. J. The turn of the screw: the bacterial flagellar motor. Cell 93, 17–20 (1998).
Lloyd, S. A. & Blair, D. F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. Mol. Biol. 266, 733–744 (1997).
Zhou, J., Lloyd, S. A. & Blair, D. F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 95, 6436–6441 (1998).
Irikura, V. M., Kihara, M., Yamaguchi, S., Sockett, H. & Macnab, R. M. Salmonella typhimurium fliG and flN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175, 802–810 (1993).
Huber, R.et al. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90° C. Arch. Microbiol. 144, 324–333 (1986).
Yamaguchi, S., Fujita, H., Ishihara, A., Aizawa, S. & Macnab, R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166, 187–193 (1986).
Hutchinson, E. G. & Thornton, J. M. PROMOTIF—a program to identify and analyze structural motifs in proteins. Prot. Sci. 5, 212–220 (1996).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Zhou, J. & Blair, D. F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273, 428–439 (1997).
Yamaguchi, S.et al. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168, 1172–1179 (1986).
Welch, M., Oosawa, K., Aizawa, S.-I. & Eisenbach, M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl Acad. Sci. USA 90, 8787–8791 (1993).
Marykwas, D. L. & Berg, H. C. Amutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 178, 1289–1294 (1996).
Tang, H., Braun, T. F. & Blair, D. F. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261, 209–221 (1996).
Garza, A. G., Biran, R., Wohlschlegel, J. & Manson, M. D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 258, 270–285 (1996).
Francis, N. R., Sosinsky, G. E., Thomas, D. & DeRosier, D. J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235, 1261–1270 (1994).
Zhao, R., Pathak, N., Jaffe, H., Reese, T. S. & Khan, S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261, 195–208 (1996).
Khan, S., Dapice, M. & Reese, T. S. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202, 575–584 (1988).
Jones, C. J., Macnab, R. M., Okino, H. & Aizawa, S.-I. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212, 377–387 (1990).
Francis, N. R., Irikura, V. M., Yamaguchi, S., DeRosier, D. J. & Macnab, R. M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89, 6304–6308 (1992).
Braun, T. F.et al. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 181, 3542–3551 (1999).
Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L. & Sweet, R. M. The crystal structure of the globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219–223 (1993).
McRee, D. E. Avisual protein crystallographic software system for X11/XView. J. Molec. Graph. 10, 44–46 (1992).
Furey, W. & Swaminathan, S. PHASES: A program package for the processing and analysis of diffraction data from macromolecules Methods Enzymol. 277, 590–620 (1997).
Brünger, A. T. X-PLOR Version 3.843, a System for X-ray Crystallography and NMR (Yale University, New Haven, (1996).
Jones, T. A., Zhou, J.-Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and locations of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 176, 307–326 (1997).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Acknowledgements
We thank H. Bellamy, A. Kolatkar, M. Mathews, J. R. Knowlton and J. D. Phillips for assistance with data collection, and R. Huber for T. maritima genomic DNA. Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of T.maritima was accomplished with support from the US Department of Energy (DoE). The SSRL Biotechnology Program is supported by the US National Institutes of Health (NIH), National Center for Research Resources, Biomedical Technology Program, and by the DoE Office of Biological and Enivronmental Research. Use of APS was supported by the DoE Basic Energy Sciences, Office of Science. Use of the BioCARS Sector 14 was supported by the NIH National Center for Research Resources. S.A.L. was partially supported by an NIH training grant. Supported by grants from the NSF (to D.F.B.) and NIH (to C.P.H.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lloyd, S., Whitby, F., Blair, D. et al. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400, 472–475 (1999). https://doi.org/10.1038/22794
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/22794
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.