Abstract
Mechanisms underlying the initiation of proliferative response are known only for a few organisms and are not understood for the medically important organisms including Entamoeba histolytica. The trans membrane kinase EhTMKB1-9 of E. histolytica is one of the early indicators of proliferation and its' expression is regulated by serum, one of the components necessary for cellular proliferation in vitro. In this study we show that bovine serum albumin (BSA) can induce EhTMKB1-9 expression in place of serum and that both follow the same mechanism. Both serum and BSA use the same promoter element and the activation process is initiated through a PI3 kinase-mediated pathway. We further show that BSA activates EhTMKB1-9 due to the lipids associated with it and that unsaturated fatty acids are responsible for activation. These results suggest that lipid molecules are ligand(s) for initiation of a signaling system that stimulates EhTMKB1-9 expression.
Similar content being viewed by others
Introduction
Amoebiasis is a major public health problem worldwide particularly in the developing world1. It is caused by infection of the protist parasite Entamoeba histolytica. The infection starts by ingestion of the cyst form of the parasite through oral route and excystation in the intestine to form trophozoites. In the majority of infected individuals trophozoites proliferate and differentiate into cysts without harming the host. Occasionally, however, trophozoites can invade tissues causing clinical symptoms. The mechanisms by which amebic trophozoites cause invasion and pathogenesis have become clear through a number of elegant studies2,3,4,5,6,7. However, there is little knowledge about basic processes, such as gene regulation, signal transduction mechanisms and proliferation of this parasite. Genome analysis has clearly shown that E. histolytica possesses extensive signaling systems. The presence of a large number of trans membrane kinase (TMK) genes suggests that E. histolytica has evolved an elaborate system for sensing the extra cellular environment8,9. Understanding the biology of TMKs may help us to learn about the mechanisms of signal perception and propagation.
All E. histolytica TMKs (EhTMKs) contain an N-terminal signal peptide, a predicted extracellular domain and a single trans membrane helix followed by a cytosolic tyrosine kinase-like domain. EhTMKs have been grouped into six distinct families based on motifs present on both the extracellular and kinase domains8. Spotted oligoarrays and real-time PCR showed that different families of EhTMKs are expressed in E. histolytica cells and that the level of expression of individual TMKs differed significantly8. The first evidence that suggested EhTMK to have a significant role in amebic biology came from studies on the EhTMKB1 family10. E. histolytica cells over expressing a truncated form of EhTMKB1-2, showed a defect in cellular proliferation. Further, evidence in support of proliferative role of B1 family of TMKs came from careful analysis of expression and by the observation that of 35 members of the EhTMKB1 family, EhTMKB1-9 is the major expressed product in mid-log phase E. histolytica cells. On down regulation of this gene a reduction in cellular proliferation was observed11. Involvement of EhTMKB1-9 in endocytosis and target cell killing was also demonstrated in the down regulated cell lines and in dominant negative cells over expressing a truncated TMKB1-9 lacking the kinase domain11. Reduced levels of the kinase also inhibited pathogenesis in animal models12. All these suggested that EhTMKB1-9 may be an important molecule involved in both cellular proliferation and virulence. Another EhTMK, EhTMKB2-96 has also been reported to be functionally important as it participates in erythrophagocytosis and pathogenesis13.
On serum starvation EhTMKB1-9 expression was down regulated in E. histolytica cells and the expression was restored within 2 h of serum replenishment. This down regulation was at the transcriptional level and the promoter region responsible for regulated expression of EhTMKB1-9 was also mapped to a region between −768 to −939 bp of the gene11. This region probably contains both the inducible and basal activities of the promoter. Therefore, this gene is one of the rare E. histolytica genes whose promoter is not close to the transcription start site.
In this report, we show that lipids associated with bovine serum albumin (BSA) can generate a signaling pathway that eventually leads to activation of serum inducible EhTMKB1-9 expression. Inducible signaling pathways are rare in protistan parasites and there are only a few examples of lipid-induced gene regulatory pathways in any system. Since serum response system has been the basis of discovery of many signaling molecules in mammalian systems, we would like to use lipid-induced EhTMKB1-9 expression as a system to decipher the mechanisms of signaling and its coupling with physiological response.
Results
Stimulation of EhTMKB1-9 expression by BSA
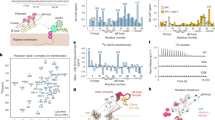
We had earlier shown that EhTMKB1-9 mRNA and protein are expressed at very low levels in serum starved E. histolytica cells and that this is reversed on serum replenishment11. Serum is an essential component in media used for E. histolytica axenic cell proliferation. In a previous report it has been shown that BSA could replace serum in the growth medium14. However it was not possible to replace BSA with any other components (such as amino acid mixture). We decided to check if the addition of BSA in place of serum could stimulate EhTMKB1-9 transcripts when added to serum-starved cells. The level of EhTMKB1-9 was estimated by northern analysis using a specific probe derived from the 5′-end of the gene11. After 24 h starvation, the level of EhTMKB1-9 mRNA was three fold lower than that of cells growing in complete medium containing serum (Fig 1a). When serum was added back to the starved cells for 2 h, the amount of transcript went up to the level observed in growing cells. Next, the starved cells were resuspended in different concentrations of BSA for 2 h and the level of EhTMKB1-9 transcript was determined. EhTMKB1-9 transcript levels increased with increasing concentration of BSA. At 10 µg/µl of BSA (equivalent to 15% of serum) the level of expressed EhTMKB1-9 was nearly equivalent to that of proliferating cells. This stimulation was not seen with mucin which was used since E. histolytica colonizes a mucin-rich environment in the human gut and also mucin is also used in some growth media15,16. The results were also confirmed by real time PCR (Fig 1b). Our data suggest that BSA may be a specific activator of EhTMKB1-9 expression.
BSA stimulates EhTMKB1-9 expression.
(a) Northern hybridization with EhTMKB1-9 specific probe was performed with RNA isolated from cells incubated in indicated medium. Serum starvation was carried out for 24 h and replenishment for 2 h as indicated. BSA bl. – Boiled BSA. Methylene blue staining of 18 s RNA was taken as loading control. Comparisons were made with respect to cells starved for 24 h and statistical significance was determined by paired t- test.*** p < 0.002; ** p < 0.005; *p < 0.01. (b) Quantitative real-time PCR was performed for measuring the level of EhTMKB1-9 and RNA polymerase II gene transcripts using cells treated under different conditions as indicated. BSA bl. – Boiled BSA. All samples were analyzed in triplicates, in three independent experiments. Values are normalized to the endogenous control (RNA Pol II) and results are expressed as percent fold change in comparison to normal proliferating cells (taken as 100%).Comparisons were made with respect to cells starved for 24 h and statistical significance was determined by paired t- test. *** p < 0.002; ** p < 0.005; *p < 0.01.
In order to show that the activation of EhTMKB1-9 gene by BSA follows a mechanism similar to that of serum replenishment of starved cells, a construct containing luciferase gene downstream of the EhTMKB1-9 promoter element (p9–939) was used. Luciferase expression responded to serum in a manner similar to that of the endogenous gene11. Serum starvation resulted in substantial reduction (about 2 fold) in luciferase activity compared to proliferating cells (Fig 2a). The activity increased by about three fold on serum replenishment as expected. With increasing amount of BSA, the luciferase activity increased and at 50 µg/µl the activity was maximal. Thereafter, the level of luciferase activity decreased even on further increase in BSA concentration. Moreover, replenishment with mucin used at 10 µg/µl did not stimulate luciferase activity. As a control, luciferase constructs containing, a) promoter of EhTMKB1-5; b) a promoter less construct and c) the lectin promoter were also tested under the same conditions and no change in luciferase activity was observed in cells containing these constructs (data not shown). These results strongly suggest that induction of EhTMKB1-9 expression by BSA may be through activation of promoter element, as observed with serum. Henceforth, we used luciferase based assay for subsequent analysis of EhTMKB1-9 gene activation.
BSA activates EhTMKB1-9 promoter and the effect of signaling inhibitors.
(a) Promoter activity was determined by using a luciferase construct containing upstream promoter element (p9–939) of EhTMKB1-9 as described before11. The cells were treated as indicated and luciferase activity was measured using a luminometer. Comparisons were made with respect to cells starved for 24 h and statistical significance were determined by paired t- test. *** p < 0.002; ** p < 0.005; *p < 0.01. (b) Luciferase based promoter assay was used to check the effect of indicated signaling inhibitors on EhTMKB1-9 promoter activation. The inhibitors were added during the time of replenishment. Comparisons were made with respect to cells treated with 15% serum and wortmannin as well as BSA and wortmannin during replenishment for 2 h and statistical significance were determined by paired t-test. *** p < 0.002; ** p < 0.005; *p < 0.01.
A number of mechanisms are likely to be involved in the activation of expression of EhTMKB1-9 by BSA or serum. Firstly, BSA/serum can act as a ligand to a hitherto unknown cell surface receptor(s), initiating a signaling cascade leading to gene activation. The other possibility is that BSA/serum provides nutritional supplementation and activation is a response towards increasing amino acid pool of the cell. In order to show that signaling pathways may be involved in the activation, experiments were carried out to check if common modulators of signaling pathways are likely to affect serum/BSA-induced EhTMKB1-9 gene expression after starvation. The results are shown in Fig. 2b. The PI3Kinase inhibitor wortmannin blocked activation of the gene even at low concentration of 25 nM, in both serum and BSA replenishment conditions (Fig 2b). However, there was no effect of rapamycin, an inhibitor of mTOR/MAP kinase pathway even at a concentration of 100 nM. Calcium signaling pathway was also not involved as there was no effect on activation of EhTMKB1-9 expression in the presence of calcium ion chelator BAPTA-AM. The results suggest that BSA is likely to initiate a PI3Kinase pathway probably through receptor molecules, leading to EhTMKB1-9 gene activation.
Subcellular localization of BSA-induced EhTMKB1-9 in E. histolytica cells
Distribution of EhTMKB1-9 molecules in cells that underwent serum starvation followed by replenishment with either serum or BSA was studied. Anti EhTMKB1-9 antibody was used in conjunction with confocal microscopy to study the distribution of EhTMKB1-9 molecules in amebic cells that had been serum starved before incubation with either serum or BSA11. High intensity of fluorescence was visible throughout the cell membrane in surface fixed, non permeabilized proliferating cells (Fig 3). After permeabilization punctate staining was also observed in the cytoplasm suggesting that the molecules are present in endocytic vesicles and the distribution is similar to that observed before11 (Fig 3). Presence of EhTMKB1-9 in newly formed phagosome was shown by us in a previous study17. On serum starvation the amount of fluorescent signal was substantially reduced. However, it was restored on serum replenishment (Fig 3). When starved cells were resuspended in BSA instead of serum, an increase in the amount of EhTMKB1-9 was observed and the pattern looked similar to that seen after serum replenishment.
Immunolocalization of EhTMKB1-9.
Confocal microscopy was employed for immunolocalization using pre-immune serum or anti-EhTMKB1-9 antibody. Both non-permeabilized and permeabilized (0.1% Triton X-100) amebic cells were stained and the cells were treated under different conditions, complete medium, serum starvation and replenishment as indicated. Bar represents 5 µm. Magnification 60X.
The stimulatory activity resides in lipids associated with BSA
Commercial preparations of BSA contain a number of lipids and lipid-like hydrophobic molecules. In order to check if the stimulation of EhTMKB1-9 expression is initiated by these components we carried out experiments with fat-free BSA (FFBSA) and lipid-like molecules that were extracted from BSA. The results are shown in Fig 4a. There was no stimulation of EhTMKB1-9 expression when FFBSA was used after serum starvation. However, when FFBSA was reconstituted with extracted lipids stimulatory activity of BSA was restored (Fig 4a). Further, we tested different combinations of commercially available lipids18,19with FFBSA in order to study specificity in this system. While unsaturated fatty acids, such as oleic and linoleic acids stimulated EhTMKB1-9 promoter no significant effect was seen when saturated fatty acids, such as stearic and palmitic acids were used in the assay (Fig 4a). Pure fatty acid, such as oleic acid in the absence of FFBSA was not effective in stimulating the EhTMKB1-9 promoter. However, in the presence of FFBSA, the level of activation of EhTMKB1-9 promoter, increased with increasing concentration of oleic acid, reaching almost saturation and was comparable to that seen in cells growing in serum containing medium (Fig 4b). Activation of EhTMKB1-9 expression by oleic acid was also inhibited by wortmannin suggesting that the signaling system that initiates this activation is lipid-induced (Fig 4c). This suggests that EhTMKB1-9 stimulatory activity in BSA resides in the lipid fraction and even a single pure lipid is capable of achieving this activation.
Effect of lipids on EhTMKB1-9 expression.
(a) Reporter luciferase activity of the stable transfectants p9–939 was used for checking the effect of lipids on EhTMKB1-9 expression. Replenishment was done using indicated compounds including lipids extracted from BSA. Fat free BSA was used as a carrier for lipids. FF BSA- Fat free BSA; Ol. Oleic acid; Ln. - Linoleic acid; St - Stearic acid; Ar.-Arachidonic acid; Pl. - Palmitic acid; Reconst. Lp.- Reconstituted lipids. The fatty acids were used at 115 µg/ml; 300 µg/ml; 166 µg/ml; 107 µg/ml; 312 µg/ml for oleic acid, linoleic acid, stearic acid, arachidonic acid and palmitic acid respectively. FF BSA were used at 10 mg/ml. Comparisons were made with respect to cells starved for 24 h and statistical significance was determined by paired - t test. *** p < 0.002; ** p < 0.005; *p < 0.01. (b) Luciferase activity of EhTMKB1-9 promoter in presence of increasing concentration of oleic acid. FF BSA - Fat Free BSA; Ol. - Oleic acid; Ol. alone – oleic acid without Fat free BSA. Oleic acid was used at 115 µg/ml (1x). FF BSA were used at 10 mg/ml. Comparisons were made with respect to cells starved for 24 h and statistical significance were determined by paired - t test.*** p < 0.002; ** p < 0.005; * p < 0.01. (c) Luciferase activity of EhTMKB1-9 promoter in presence of Oleic acid and inhibitor wortmannin. FF BSA - Fat free BSA; wort – wortmannin; Ol.- Oleic acid; Oleic acid was used at 460 µg/ml(4x). FF BSA were used at 10 mg/ml. Comparisons were made with respect to cells treated with Fat free BSA and oleic acid with and without wortmannin during replenishment and statistical significance was determined by paired-t test. *** p < 0.002; ** p < 0.005; * p < 0.01.
Discussion
The most worked out systems to study signaling pathways have been a few bacterial species and eukaryotic model systems, such as Drosophila, C. elegans and mammals. In general, cell surface interaction of growth factors with their cognate receptors starts a cascade of signaling pathways involving protein kinases20,21. During the activation process different genes get switched on/off at different times depending on their role in cell proliferation. For example, the immediate early gene c-fos gets switched on within minutes of the growth factor binding to its receptor22. In the primitive parasitic protist E. histolytica EhTMKB1-9 was shown to be one of the early serum response genes11 and its expression was low in serum starved cells (non proliferating). However, on serum replenishment the expression was induced within 30 min. A significant decrease in cellular proliferation was observed in cell lines expressing either EhTMKB1-9 without kinase domain, or EhTMKB1-9 antisense RNA11 suggesting that this transmembrane kinase is part of the signaling pathway that is involved in cell multiplication. In this report, we provide evidence to suggest that lipids associated with BSA are the ligand(s) that stimulate the PI3Kinase based signaling system leading to EhTMKB1-9 gene activation. This is one of the first signaling ligands that have been described for any protist parasite and one of the few systems where lipids have been reported as signal initiating ligands.
The results presented here clearly show that activation of EhTMKB1-9 by BSA after serum starvation follows the same mechanism as that observed with serum. The level of activation and time kinetics were found to be similar in both cases. The stimulation of transcription by serum and BSA involved activation of promoter elements and was blocked by the PI3kinase inhibitor wortmannin. Therefore transcriptional activation is likely to be a receptor mediated process rather than a nutritional response through PI3K pathway which has been shown to be involved in proliferation in other systems20. There is a significant cross talk between PI3K and MAPK pathways leading to activation of AKT and phosphorylation of ERK20. E. histolytica genome encodes a number of genes that participate in phosphoinositide metabolism. Out of 12 phosphoinositide-binding FYVE domain containing proteins, 11 also have RhoGEF/DH domain suggesting their involvement in regulating cytoskeleton dynamics23. One of these proteins EhFP4 was shown to participate in phagocytosis through binding Rho/Rac small GTPase. Though details of phosphoinositide metabolism and their biological role are not yet known in E. histolytica it appears that these molecules may have a major function in signaling processes. We plan to use antisense-based down regulation as a strategy to identify the gene(s) belonging to phosphoinositide pathway that are involved in EhTMKB1 transcriptional activation.
Our results show that BSA dependent activation of EhTMKB1-9 gene is actually due to the lipids associated with BSA, as the lipid fraction, but not the fat-free BSA could stimulate gene expression. BSA contains mainly palmitic acid, oleic acid, stearic acid, linoleic acid and arachidonic acid; apart from a number of minor components19,24. Lipid composition of BSA reflects that of serum18,24. Since the unsaturated fatty acids (oleic and linoleic acids) could and the saturated acids (stearic and palmitic acids) could not stimulate transcription from EhTMKB1-9 promoter, it appears that the lipid recognition system may have some specificity. These results indicate a specific signaling system through PI3K that is involved in initiating transcription activation of one of the early proliferation related gene EhTMKB1-9 and rule out the role of BSA as a source of amino acids.
Several reports have suggested the role of fatty acids in regulating the expression of many genes involved in lipid metabolism25,29 and modulating the activity of signaling molecules, such as phosphatidylinositol 3-kinase/AKT, NF-kappaB26,27via the Toll like receptor family. Moreover, Toll like receptors can be either activated by saturated fatty acids or suppressed by unsaturated fatty acids suggesting that lipids can be ligands for TLR activation25,28,29. Free fatty acid receptors have been identified in pancreatic beta cells and these have been implicated in regulation of insulin secretion30. Interestingly, there was no effect when lipids associated with BSA were used in this system unlike that observed by us. Though these receptors are conserved in a large number of organisms we could not find any homolog in the E. histolytica genome. It is likely that amebic receptors may be different compared to those in higher organisms. In the context of amoebiasis the role of diet and nutritional status of the host in the progress of disease has been suggested in studies with animal models31. Our results provide a clue to the mechanism by which diet may affect cellular proliferation and thereby disease progression, in E. histolytica infection.
In conclusion, we believe that lipid-induced EhTMKB1-9 expression is an immediate early response of growth-stressed E. histolytica to initiate proliferation. Though we do not know the function of this kinase, it is likely that it may be involved in further signal transduction, being a cell surface molecule containing a functional kinase domain11. For the first time we have described a defined inducible system for gene activation leading to cellular proliferation in E. histolytica. This pathway may be a good drug target for developing therapeutics against amoebiasis.
Methods
Strains and cell culture
All experiments were carried out with E. histolytica strain HM-1:IMSS clone 6. The cells were maintained and grown in TYI-33 medium supplemented with 15% adult bovine serum, 1X diamond's vitamin mix and antibiotics (0.3 units/µl penicillin and 0.25 µg/µl streptomycin) at 35.5°C32. To achieve serum starvation condition, media from mid log phase grown E. histolytica cells was replaced with TYI-33 medium containing 0.5% adult bovine serum for indicated period of time. Replenishment was achieved by decanting the media after 24 h of serum starvation and replacing with media containing indicated compounds for 2 h. G-418 (Sigma) was added at 10 µg/ml for maintaining the transfected cell lines.
Northern hybridization
Total RNA was purified using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA samples (30 µg) were resolved in formaldehyde agarose in gel running buffer [0.1 M MOPS (pH 7.0), 40 mM sodium acetate, 5 mM EDTA (pH 8.0)] and 37% formaldehyde at 4 V/cm. The RNA was transferred on to GeneScreen plus (NEN) nylon membranes. Hybridization and washing conditions for RNA blots were as per manufacturer's protocol.
Quantitative Real Time (qRT-PCR)
Real time PCR efficiencies for each gene were calculated from the slope, according to the established equation E = 10[−1/slope] using genomic DNA as template (serial 1∶10 fold dilutions) and were found around 1.96±0.0633. Two µg total RNA (DNase I treated) was reverse transcribed using random hexamers into cDNA by Superscript III reverse transcriptase (Invitrogen). Real time quantitative PCR was performed in 7500 Real Time PCR System (Applied Biosystems) using SYBR green PCR Master Mix, 2 pmol of forward and reverse primers and 2 µl of cDNA (serial 1∶10 fold dilution). EhTMKB1 members and the RNA Pol II (control gene) were amplified in parallel. The conditions were predenaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 58°C for 1 min followed by a dissociation stage at 95°C for 15 sec and 58°C for 1 min. Cycle threshold values (Ct) were analyzed by the SDS1.4 software (Applied Biosystems) and all samples were analyzed in triplicates in three independent experiments. Reactions without cDNA were used as no template control and no RT controls were also set up to rule out genomic DNA contamination. Relative quantification of EhTMKB1expression was determined using the comparative Ct method (ABI Prism 7500, SDS User Bulletin; Applied Biosystems).
Luciferase Assay
The procedure was done as described previously34. Briefly, stably transfected trophozoites, maintained in TYI-S-33 medium supplemented with 10 µg/µl G-418, were chilled on ice, harvested and washed once in PBS (pH 7.4) and lysed in 200 µl of reporter lysis buffer (Promega) with the addition of protease inhibitors E64-C and leupeptin. Lysates were frozen overnight at −80°C. After thawing on ice for 10 min, cellular debris was pelleted and the samples were allowed to warm to room temperature. Luciferase activity was measured according to the manufacturer's instructions (Promega) using a Turner Luminometer (model TD-20E). Luciferase activity per µg of protein was calculated as a measure of reporter gene expression.
Immunofluoroscence staining
Immunofluorescence staining was carried out as described before35. Briefly E. histolytica cells resuspended in TYI-33 medium were transferred onto acetone-cleaned coverslips placed in a petri dish and allowed to adhere for 10 min at 35.5°C. The culture medium was removed and cells were fixed with 3.7% pre-warmed paraformaldehyde (PFA) for 30 min. After fixation, the cells were permeabilized with 0.1% Triton X-100/PBS for 1 min. This step was omitted for non-permeabilized cells. The fixed cells were then washed with PBS and quenched for 30 min in PBS containing 50 mMNH4Cl. The coverslips were blocked with 1% BSA/PBS for 30 min, followed by incubation with primary antibody at 37°C for 1 h. The coverslips were washed with PBS followed by 1% BSA/PBS before incubation with secondary antibody of 30 min at 37°C. Antibody dilutions used were: anti-EhTMKB1-9 at 1∶20, anti-rabbit Alexa 488 (Molecular Probes) at 1∶300. The preparations were further washed with PBS and mounted on a glass slide using DABCO [1, 4-diazbicyclo (2,2,2) octane (Sigma) 10 µg/µl in 80% glycerol]. The edges of the coverslip were sealed with nail-paint to avoid drying. Confocal images were visualized using an Olympus Fluoview FV1000 laser scanning microscope.
Reconstitution of lipid-associated BSA and lipids extracted from intact BSA
Dried lipid precipitates were prepared by evaporating the organic solvent from an appropriate amount of lipid stock solutions in ethanol or chloroform under nitrogen-gas stream aseptically.
To prepare the reconstituted lipid-associated BSA, fat-free BSA stock solution (10 µg/µl) prepared in TYI-33 medium was added to the dried lipid precipitate and the mixture was sonicated twice for 1 min until the dried precipitate got dissolved. Total lipids from 1.3 g of intact BSA were extracted by the method of Bligh and Dyer36.
References
Bercu, T. E., Petri, W. A. & Behm, J. W. Amebic colitis: new insights into pathogenesis and treatment. Curr Gastroenterol Rep. 9, 429–33 (2007).
Petri, W. A., Jr et al. Recognition of the galactose- or N-acetylgalactosamine-binding lectin of Entamoeba histolytica by human immune sera. Infect Immun. 55, 2327–31 (1987).
Clark, C. G. et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol. 65, 51–190 (2007).
Stanley, S. L., Jr, Zhang, T., Rubin, D. & Li, E. Role of the Entamoeba histolytica cysteine proteinase in amebic liver abscess formation in severe combined immunodeficient mice. Infect Immun. 63, 1587–90 (1995).
Ankri, S. et al. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol Microbiol. 33, 327–37 (1999).
Ankri, S., Stolarsky, T., Bracha, R., Padilla-Vaca, F. & Mirelman, D. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect Immun. 67, 421–2 (1999).
Bracha, R., Nuchamowitz, Y. & Mirelman, D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell 2, 295–305 (2003).
Beck, D. L. et al. Identification and gene expression analysis of a large family of transmembrane kinases related to the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell 4, 722–32 (2005).
Loftus, B. et al. The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–8 (2005).
Mehra, A., Fredrick, J., Petri, W. A., Jr, Bhattacharya, S. & Bhattacharya, A. Expression and function of a family of transmembrane kinases from the protozoan parasite Entamoeba histolytica. Infect Immun. 74, 5341–51 (2006).
Shrimal, S., Bhattacharya, S. & Bhattacharya, A. Serum-dependent selective expression of EhTMKB1-9, a member of Entamoeba histolytica B1 family of transmembrane kinases. PLoS Pathog. 6, e1000929 (2010).
Abhyankar, M. A., Shrimal, S., Gilchrist, C., Bhattacharya, A. & Petri, Jr, W. A. The Entamoeba histolytica serum-inducible transmembrane kinase EhTMKB1-9 is involved in intestinal amebiasis. Intern. J. Parasitol. Drugs & Drug Resistance. (2012) In press.
Boettner, D. R. et al. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 4, e8 (2008).
Diamond, L. S. & Cunnick, C. C. A serum-free, partly defined medium, PDM-805, for axenic cultivation of Entamoeba histolytica Schaudinn, 1903 and other Entamoeba. J Protozool. 38, 211–6 (1991).
Lidell, M. E., Moncada, D. M., Chadee, K. & Hansson, G. C. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci U S A. 103, 9298–303 (2006).
Dolkart, R. E. & Halpern, B. A new monophasic medium for the cultivation of Entamoeba histolytica. Am J Trop Med Hyg. 7, 595–6 (1958).
Somlata, Bhattacharya, S., Bhattacharya, A. A C2 domain protein kinase initiates phagocytosis in the protozoan parasite Entamoeba histolytica. Nat Commun. 2, 230 (2011)
Saifer, A. & Goldman, L. The free fatty acids bound to human serum albumin. J Lipid Res. 2, 268–270 (1961).
Hjartaker, A., Lund, E. & Bjerve, K. S. Serum phospholipid fatty acid composition and habitual intake of marine foods registered by a semi-quantitative food frequency questionnaire. Eur J Clin Nutr. 51, 736–42 (1997).
Zhang, W. & Liu, H. T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002).
Seger, R. & Krebs, E. G. The MAPK signaling cascade. Faseb J. 9, 726–35 (1995).
Greenberg, M. E. & Ziff, E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311, 433–8 (1984).
Nakada-Tsukui, K., Okada, H., Mitra, B. N. Nozaki, T. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell Microbiol. 10, 1471–91 (2009).
Chen, R. F. Removal of fatty acids from serum albumin by charcoal treatment. J Biol Chem. 242, 173–81 (1967).
Fessler, M. B., Rudel, L. L. & Brown, J. M. Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol. 20, 379–85 (2009).
Lee, J. Y. et al. Reciprocal modulation of Toll-like receptor-4 signaling pathways involving MyD88 and phosphatidylinositol 3-kinase/AKT by saturated and polyunsaturated fatty acids. J Biol Chem. 278, 37041–51 (2003).
Suganami, T. et al. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler Thromb Vasc Biol. 27, 84–91 (2007).
Lee, J. Y. et al. Differential modulation of Toll-like receptors by fatty acids: preferential inhibition by n-3 polyunsaturated fatty acids. J Lipid Res. 44, (479–86) (2003).
Lee, J. Y. et al. Saturated fatty acid activates but polyunsaturated fatty acid inhibits Toll-like receptor 2 dimerized with Toll-like receptor 6 or 1. J Biol Chem. 279, 16971–9 (2004).
Itoh, Y. et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature 422, 173–6 (2003).
Diamond, L. S. Amebiasis: nutritional implications. Rev Infect Dis. 4, 843–50 (1982).
Diamond, L. S., Harlow, D. R. & Cunnick, C. C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 72, 431–2 (1978).
Pfaffl, M. W. (2001) A new mathematical model for relative quantification in realtime RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Ramakrishnan, G., Vines, R. R., Mann, B. J. & Petri, W. A., Jr A tetracycline inducible gene expression system in Entamoeba histolytica. Mol Biochem Parasitol. 84, 93–100 (1997).
Sahoo, N. et al. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J Cell Sci. 117 3625–3634 (2004).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 37, 911–7 (1959).
Acknowledgements
The authors thank Department of Biotechnology and Department of Science & Technology for financial support and JC Bose fellowship. A donation from BNP Paribas is gratefully acknowledged. The authors also thank Dr. Pritam Mukherji of School of Physical Sciences for providing chemistry laboratory support.
Author information
Authors and Affiliations
Contributions
SS, AS, AB, SB wrote the main manuscript text and SS and AS prepared the figures 1–4. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Shrimal, S., Saha, A., Bhattacharya, S. et al. Lipids induce expression of serum-responsive transmembrane kinase EhTMKB1-9 in an early branching eukaryote Entamoeba histolytica. Sci Rep 2, 333 (2012). https://doi.org/10.1038/srep00333
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00333
This article is cited by
-
Identification and Characterization of Entamoeba histolytica Choline Kinase
Acta Parasitologica (2024)
-
Computational Modeling of the Activity of Metronidazole against EhGα1 of Entamoeba histolytica Enhanced by its Copper and Zinc Complexes
Chemistry Africa (2021)
-
Transcriptomic analysis reveals novel downstream regulatory motifs and highly transcribed virulence factor genes of Entamoeba histolytica
BMC Genomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.