Abstract
Mitochondrial DNA sequences are frequently transferred into the nuclear genome, generating nuclear mitochondrial DNA sequences (NUMTs). Here, we analysed, for the first time, NUMTs in the domestic yak genome. We obtained 499 alignment matches covering 340.2 kbp of the yak nuclear genome. After a merging step, we identified 167 NUMT regions with a total length of ~ 503 kbp, representing 0.02% of the nuclear genome. We discovered copies of all mitochondrial regions and found that most NUMT regions are intergenic or intronic and mostly untranscribed. 98 different NUMT regions from domestic yak showed high homology with cow and/or wild yak genomes, suggesting selection or hybridization between domestic/wild yak and cow. To rule out the possibility that the identified NUMTs could be artifacts of the domestic yak genome assembly, we validated experimentally five NUMT regions by PCR amplification. As NUMT regions show high similarity to the mitochondrial genome can potentially pose a risk to domestic yak DNA mitochondrial studies, special care is therefore needed to select primers for PCR amplification of mitochondrial DNA sequences.
Similar content being viewed by others
Introduction
The evolution and function of eukaryotic genomes have strongly been influenced by the integration of mitochondrial DNA sequences into the nuclear genome1. Because of their homology, these nuclear mitochondrial DNA sequences, known as NUMTs, can compromise mitochondrial DNA studies if they are not taken into account. Indeed, the inadvertent analysis of NUMTs as mitogenomic sequences can lead to misleading results in the diagnosis of mitochondrial diseases, phylogenetic reconstructions, population studies and DNA barcoding analyses1,2. For example, Yao and collaborators revisited mitochondrial DNA sequence variations which were caused by accidental amplification of NUMTs2. They described several cases, included one NUMT sharing homologies with mitochondrial ND5 gene and generating a false mitochondrial variant believed to be responsible for hearing loss. It is therefore important to identify nuclear sequences of mitochondrial origin.
To date, a catalogue of NUMTs has been established for a large number of different avian and mammalian species3,4,5,6,7,8,9 including several ruminant species10,11,12, but not yet in the domestic yak (Bos grunniens). For example, we identified 166 bovine and 390 ovine NUMT regions, representing approximately 0.02% of the nuclear genome in each species10,11.
The domestic yak is a very important species for the inhabitants of the Qinghai-Tibetan Plateau and the Himalayan main range, providing food (milk and meat), fibre, fertiliser and carrying heavy loads. As a result, a reference genome sequence of the domestic yak has been available since 201213 and several studies have been conducted to describe the genetic diversity of domestic yak populations based on mitochondrial DNA sequences14,15,16.
Based on the analysis of whole-genome sequences of 59 domestic and 13 wild yaks, Qiu and colleagues estimated that domestication began about 7300 years ago17. Genetic introgression between domestic yaks and cattle has been reported18,19,20,21,22,23. NUMTs in domestic yak can possibly affect the previous domestication and introgression studies.
In this paper, we present the first study of NUMTs in the domestic yak genome. This study will therefore allow a more accurate analysis of the mitogenome of this yak species.
Material and methods
Data sources
The mitochondrial reference sequence and the latest BosGru3.0 domestic yak reference genome sequence24 were obtained from Genbank (assembly accessions: NC_006380.3 and GCA_005887515.1, 10th June 2019 respectively). Reference genome sequences of wild yak (Bos mutus) BosGru_v2.017 and cow genome (Bos taurus) ARS-UCD1.3 (Genbank assembly accessions: GCA_000298355.1, 9th January 2013 and GCA_002263795.4, 1st July 2023, respectively) were also retrieved from Genbank.
Genome-wide identification of NUMT regions
To deal with the fact that the mitochondrial genome is circular, we perform alignments on the nuclear genome of the domestic yak using two different linearised sequences of the mitogenome: (1) a standard linearisation starting at position 1 and ending at position 16,323; and (2) a shifted linearisation starting arbitrarily at position 5001 and ending at position 5000. This allowed the identification of nuclear genome sequences that overlapped both ends of the standard linear mitochondrial genome sequence. These two linearised mitogenome sequences were aligned to the domestic yak genome sequence using BLASTN25, following the procedure described by Calabrese et al.3. However, an e-value threshold of 10–4, comparable to other studies of NUMTs1, was applied. NUMTs that were not more than 10 kbp apart on the nuclear genome were merged into one NUMT region10,11,26.
Characterization of NUMT regions
Gene annotation of the domestic yak genome was retrieved from ENSEMBL (release 106). To explore if identified NUMT regions are transcribed, we performed blastn alignments with Bos grunniens sequences from GenBank refseq_rna and EST databases. NUMTs detected on unplaced scaffolds have been excluded. In order to better investigate the possible transcription, we use ORF Finder27 to predict open reading frames (ORFs) in the sequence of NUMT regions that have been identified. We further annotated the predicted full-length ORFs using the CD search tool28.
Experimental validation of NUMT regions
PCR primers were designed within the flanking regions of the NUMT regions using primer-blast 29. In addition, each primer sequence was aligned using blastn onto the domestic yak reference genome sequence and the mitochondrial genome sequence in order to verify its nuclear specificity. Primers were purchased from Integrated DNA Technologies and sequences can be found in Supplementary material—Table S1. Polymerase chain reactions were done as previously described10. Briefly, PCRs were performed in 10 μl, using 50 ng of genomic DNA from a male domestic yak of Mongolian origins, 1 U GoTaq DNA polymerase (Promega), 1X PCR buffer, 1.5 mM MgCl2, 200 μM of each dNTP and 1.0 μM of each PCR primer. The following touchdown cycling protocol was used: 95 °C for 2 min followed by 13 cycles of 95 °C for 1 min, 1 min of annealing (the annealing temperature was progressively lowered from 68 to 56 °C in steps of 1 °C every cycle) and 72 °C for 2 min. These initial cycles were followed by 30 cycles of 95 °C for 1 min, 55 °C for 1 min and 72 °C for 2 min, and a final extension step at 72 °C for 5 min. PCR products were analysed by electrophoresis on an 1% agarose gel to verify the expected length of amplicons. The nucleotide sequences of the amplicons were subsequently determined using Sanger sequencing (Eurofins Genomics). All sequences were visually inspected using Chromas (Technelysium) and then aligned to the BosGru3.0 domestic yak reference genome sequence using blastn.
Results and discussion
NUMT catalogue in domestic yak
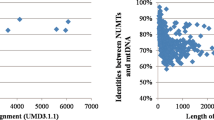
Alignment matches between the domestic yak genome sequence and the two linearised mitogenome sequences were detected in all chromosomes, including the Y chromosome (Fig. 1). We identified 499 NUMTs (alignment matches), with similarity between nuclear and mitochondrial sequences ranging between 62.9 and 100%. The total length of the alignment was 340.2 kbp, with alignment matches ranging from 31 to 11,900 bp. About 13% of the alignment matches between mitochondrial and nuclear sequences show sequence similarity higher than 85%. For example, a contiguous alignment match of 3,018 bp with similarity of 89.6% was found between region 98,761,848–98,764,866 of chromosome 3 and the mitogenome (position 7520–10,540) (Fig. 2). Mitochondrial regions present in each NUMT were defined based on the positions of the alignment matches in the mitogenome. All mitochondrial genes had alignment matches with NUMTs, however distinct numbers of copies were detected (Fig. 3). Some of the most occurring fragments of the mitogenome in the nuclear genome included the D-loop control region and neighbouring genes (i.e. l-rRNA, ND1, CytB). While the mechanisms of mitochondrial DNA integration into the nuclear genome are not yet fully understood, interestingly, Rothfuss et al.30 have shown in a human neuroblastoma cell line that the D-loop region is more prone to breakage than the rest of the mitochondrial genome. Moreover, Doynova et al.31 have showed that the D-loop region has a greater propensity for interaction with the nuclear genome, in cultured human and mouse cells. All these results suggest that the high number of nuclear sequences originating from the mitochondrial D-loop region may be arise from a higher number of direct integrations into the nuclear genome.
Dot plot representing the alignment of a large nuclear mitochondrial DNA sequence (Y.3.84) with the domestic yak mitogenome. Sequences of the mitochondrial genome and of the sequence of the NUMT region are plotted on X axis and Y axis respectively. The positions indicated in the axes of the dot plot start at 1 and go to the complete length of the sequence. Therefore, dot plot representation is not on the same scale for the X and Y axes.
Nuclear copies of mitochondrial origin can be highly modified after integration by insertions and deletions. Consequently, NUMTs that were no more than 10 kbp apart on the nuclear genome were merged into one NUMT region. After this merging step, we identified a total of 167 NUMT regions ranging from 31 to 35,446 bp (Supplementary material—Table S2 and Fig. S1). We identified 35 NUMT regions with more than 5000 bp, representing ~ 21% of the total number of NUMTs regions and around 80% of the NUMT region total length. By contrast, NUMT regions smaller than 300 bp comprise ~ 45% of the total NUMT regions length, but represent only ~ 2% of the NUMT region total length. The total length of the NUMT regions reached 502.6 kbp, representing 0.02% of the domestic yak nuclear genome. This represents almost a twofold increase of the total length detected after merging co-linear NUMTs. Interestingly we also found the same proportion of 0.02% of the genome including NUMT regions in cattle10 and sheep11.
NUMT conservation among cattle, domestic and wild yaks
To investigate the conservation of the NUMT regions identified in the domestic yak, their sequences were aligned, after masking repeats, to the wild yak (Bos mutus) and cow genomes. 36,964 and 206,374 hits (e-value threshold of 10–4) were obtained with the wild yak and cow genomes, respectively. Only the 35 bp long NUMT Y.17.344 did not match any species. 82 different NUMT regions from the domestic yak showed high homology (identity percentage > 90%) with the wild yak and cow genomes. 16 NUMT regions were shared only by both yak species. Interestingly, 32 NUMT regions were highly conserved (> 99% identity) among the three species (Supplementary Material—Table S3). This high conservation could be due to selection or hybridisation between domestic/wild yak and cow. Indeed, hybridisation between domestic and wild yaks or between cattle and domestic yaks has been reported32. Domestic yak and cattle (Bos taurus) are capable of producing offspring. F1 hybrid males are sterile, but F1 hybrid females are fertile and can be backcrossed to cattle or yak bulls18. Several studies have described the introgression of bovine sequences into domestic yak genomes18,19,20,21,22,23.
Characterization and in silico analysis of the expression of nuclear copies of mitochondrial genes
We compared the location of the 167 NUMT regions using the latest gene annotation of the domestic yak. NUMTs are mostly located in non-genic regions. We identified only 48 NUMT regions overlapping with 46 genes (45 NUMT regions) and 3 pseudogenes (3 NUMT regions; Supplementary material—Table S4). We found that 31 NUMT regions overlapping with genes are located in introns. For example, NUMT region Y1.7 is located within intron 3 of TNFSF10. All remaining NUMTs contains 5′ or 3′ untranslated regions, including sometime part of the first exon. In addition, one NUMT (Y.10.193), located on chromosome 10 and 10.7 kb long encompasses the full sequence of a novel single-exon gene (ENSBGRG00000022824) encoding a 112 amino-acid protein. This gene is conserved in several mammalian species, including American bison and zebu.
To investigate if these mitochondrial nuclear sequences are transcribed, we performed blastn alignments with refseq_rna and EST databases from GenBank of Bos grunniens. NUMTs found on unplaced scaffolds were excluded. We identified significant blast hits (e value threshold of 10−4 and identity of at least 98%) with refseq_rna sequences for 12 NUMT regions. These sequences match 12 predicted non-coding RNA sequences, although some of these sequences are transcript isoforms of the same gene. (Supplementary material—Table S5). Interestingly none of these refseq_rna sequences share significant homology to the mitochondrial genome, however at least six of these refseq_rna sequence contain domains associated to mitochondrial proteins (e.g. cytochrome P450 domain). We did not find significant matches between the sequences of NUMTs and ESTs. However, only 74 ESTs from domestic yak are currently deposited in NCBI databases, all generated from a single mammary gland sample. The results of these sequence comparisons suggest the possibility that some of the NUMT regions we identified contain elements that might be transcribed.
In order to further examine the possible transcription of the identified NUMTs, we predicted with ORF Finder open reading frames located within the sequences of each NUMT region. 7220 ORFs were predicted, ranging from 29 to 1286 amino acids. We found 258 partial ORFs, containing a start codon but without a stop codon, among these ORFs. The remaining part of those predicted ORFs may be found in the nuclear sequences flanking these NUMT regions. We found 110 different conserved protein domains, within 175 ORFs from 61 NUMT regions. As expected, most of these protein domains are shared with proteins encoded by the mitochondrial genome (e.g. CYTB domain).
Experimental validation of NUMT regions
In order to exclude the possibility that the identified NUMTs could be artefacts of the latest domestic yak genome assembly, we randomly selected five NUMT regions located on five different chromosomes for experimental validation. All five amplicons were of the expected length supporting the absence of genome assembly artefacts (Fig. 4). The nucleotide sequence of each amplicon was subsequently partially determined using Sanger sequencing (Eurofins Genomics). blastn alignment of the amplicon sequences to the domestic yak reference genome sequence confirmed the presence of these NUMT regions.
Conclusion
Our study provides the first comprehensive description of NUMTs in the domestic yak genome. Interestingly we found 34 NUMTs highly conserved among domestic yak, wild yak and cattle, suggesting hybridization between these species or that these regions are under selection. We found several NUMT regions showing high similarity to the mitochondrial genome that could potentially pose a risk to mitochondrial studies. For example, we discovered that a portion of NUMT region Y.26.469 located on chromosome 26 shares > 97% identity with a mitochondrial sequence containing the D-loop control region, which is often used in genetic diversity and phylogenetic studies33.
Therefore special care should be taken when selecting primers for PCR amplification of mitochondrial DNA.
Data availability
Analyses used as raw data publicly available data (i.e. the genome reference sequences of the domestic yak, of the wild yak and of the cow, in addition to the domestic yak mitochondrial sequence, see “Material and method” section). All data supporting the findings of this study are available as part of the article and no additional source data are required.
References
Hazkani-Covo, E., Zeller, R. M. & Martin, W. Molecular poltergeists: Mitochondrial DNA copies (NUMTs) in sequenced nuclear genomes. PLoS Genet. 6, e1000834 (2010).
Yao, Y.-G., Kong, Q.-P., Salas, A. & Bandelt, H.-J. Pseudomitochondrial genome haunts disease studies. J. Med. Genet. 45, 769–772 (2008).
Calabrese, F. M. et al. NumtS colonization in mammalian genomes. Sci. Rep. 7, 16357 (2017).
Hazkani-Covo, E. & Martin, W. F. Quantifying the number of independent organelle DNA insertions in genome evolution and human health. Genome Biol. Evol. 9, 1190–1203 (2017).
Zhang, G. et al. Genomic landscape of mitochondrial DNA insertions in 23 bat genomes: Characteristics, loci, phylogeny, and polymorphism. Integr. Zool. 17, 890–903 (2022).
Qu, H., Ma, F. & Li, Q. Comparative analysis of mitochondrial fragments transferred to the nucleus in vertebrate. J. Genet. Genomics 35, 485–490 (2008).
Liang, B. et al. Comparative genomics reveals a burst of homoplasy-free Numt insertions. Mol. Biol. Evol. 35, 2060–2064 (2018).
Torres, L., Bretagnolle, V. & Pante, E. Translocation of mitochondrial DNA into the nuclear genome blurs phylogeographic and conservation genetic studies in seabirds. R. Soc. Open Sci. 9, 211888 (2023).
Baltazar-Soares, M., Karell, P., Wright, D., Nilsson, J.A. & Brommer, J.E. Bringing to light nuclear-mitochondrial insertions in the genomes of nocturnal predatory birds. Mol. Phylogenet. Evol. 181, 107722 (2022).
Tramontin, G. E. et al. Survey of mitochondrial sequences integrated into the bovine nuclear genome. Sci. Rep. 10, 2077 (2020).
Féménia, M., Charles, M., Boulling, A. & Rocha, D. Identification and characterisation of mitochondrial sequences integrated into the ovine nuclear genome. Anim. Genet. 52, 556–559 (2021).
Hassanin, A., Bonillo, C., Nguyen, B. X. & Cruaud, C. Comparisons between mitochondrial genomes of domestic goat (Capra hircus) reveal the presence of numts and multiple sequencing errors. Mitochondr. DNA 21, 68–76 (2010).
Qiu, Q. et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–969 (2012).
Guo, S. et al. Origin of mitochondrial DNA diversity of domestic yaks. BMC Evol. Biol. 6, 73 (2006).
Lai, S. J., Chen, S. Y., Liu, Y. P. & Yao, Y. G. Mitochondrial DNA sequence diversity and origin of Chinese domestic yak. Anim. Genet. 38, 77–80 (2017).
Wang, X. et al. Mitogenomic diversity and phylogeny analysis of yak (Bos grunniens). BMC Genomics 22, 325 (2021).
Qiu, Q. et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 6, 10283 (2015).
Tumennasan, K., et al. Fertility investigations in the F1 hybrid and backcross progeny of cattle (Bos taurus) and yak (B. grunniens) in Mongolia. Cytogenet. Cell Genet. 78, 69–73 (1997).
Qi, X. B. et al. Assessment of cattle genetic introgression into domestic yak populations using mitochondrial and microsatellite DNA markers. Anim. Genet. 41, 242–252 (2010).
Medugorac, I. et al. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nat. Genet. 49, 470–475 (2017).
Wu, D. D. et al. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat. Ecol. Evol. 2, 1139–1145 (2018).
Zhang, K., Lenstra, J. A., Zhang, S., Liu, W. & Liu, J. Evolution and domestication of the Bovini species. Anim. Genet. 51, 637–657 (2020).
Li, R. et al. Whole-genome analysis deciphers population structure and genetic introgression among bovine species. Front. Genet. 13, 847492 (2022).
Zhang, S. et al. Structural variants selected during yak domestication inferred from long-read whole-genome sequencing. Mol. Biol. Evol. 38, 3676–3680 (2021).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Tsuji, J., Frith, M. C., Tomii, K. & Horton, P. Mammalian NUMT insertion is non-random. Nucleic Acids Res. 18, 9073–9088 (2012).
Rombel, I. T., Sykes, K. F., Rayner, S. & Johnston, S. A. ORF-FINDER: A vector for high-throughput gene identification. Gene 282, 33–41 (2002).
Marchler-Bauer, A. & Bryant, S. H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 (2004).
Ye, J. et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 134 (2012).
Rothfuss, O., Gasser, T. & Patenge, N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 38, e24 (2010).
Doynova, M. D. et al. Interactions between mitochondrial and nuclear DNA in mammalian cells are non-random. Mitochondrion 30, 87–96 (2016).
Wiener, G., Jianlin, H., & Ruijun, L. The yak. In The Regional Office for Asia and the Pacific. (Food and Agriculture Organization of the United Nations, 2003).
Dymova, M. A. et al. Mitochondrial DNA analysis of ancient sheep from Altai. Anim. Genet. 48, 615–618 (2017).
Acknowledgements
We are grateful to Cécile Grohs (INRAE, Jouy-en-Josas) for providing the male domestic yak DNA sample. The work was supported by the National Research Institute for Agriculture, Food and Environment (INRAE).
Author information
Authors and Affiliations
Contributions
D.R. planned and coordinated the study, performed some bioinformatics analyses, analysed the results and wrote the manuscript. C.P. performed the in silico identification of NUMTs in domestic yak. M.C. did the prediction of potential ORFs within the NUMT regions. M.P. and M.F. validated some NUMT regions by PCR. A.B. and A.C. helped with laboratory experiments. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Poncet, M., Féménia, M., Pierre, C. et al. Nuclear sequences of mitochondrial origin in domestic yak. Sci Rep 14, 10217 (2024). https://doi.org/10.1038/s41598-024-61147-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-61147-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.