Abstract
Cyclin F (encoded by CCNF gene) has been reported to be implicated in the pathobiology of several human cancers. However, its potential clinical significance in clear cell renal cell carcinoma (ccRCC) remains unknown. The present study aimed to evaluate the potential significance of cyclin F, assessed by immunohistochemical (IHC) staining and molecular (bioinformatics) techniques, as a prognostic marker in ccRCC in relation to clinicopathological features and outcomes. IHC staining was performed using two independent ccRCC tissue array cohorts, herein called tissue macroarray (TMA)_1 and tissue microarray (TMA)_2, composed of 108 ccRCCs and 37 histologically normal tissues adjacent to the tumor (NAT) and 192 ccRCCs and 16 normal kidney samples, respectively. The mRNA expression data were obtained from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) public datasets, followed by bioinformatics analysis of biological mechanisms underlying prognosis. The relationship between immune cell infiltration level and CCNF expression in ccRCC was investigated using the Tumor Immune Estimation Resource 2.0 (TIMER2) and Gene Expression Profiling Interactive Analysis 2 (GEPIA2). Cyclin F expression was significantly elevated in ccRCC lesions compared to both NAT and normal renal tissues. Likewise, CCNF mRNA was markedly increased in ccRCCs relative to non-cancerous tissues. In all analyzed cohorts, tumors with features of more aggressive behavior were more likely to display cyclin F/CCNF-high expression than low. Furthermore, patients with high cyclin F/CCNF expression had shorter overall survival (OS) times than those with low expression. In addition, multivariable analysis revealed that cyclin F/CCNF-high expression was an independent prognostic factor for poor OS in ccRCC. Enrichment analysis for mechanistically relevant processes showed that CCNF and its highly correlated genes initiate the signaling pathways that eventually result in uncontrolled cell proliferation. CCNF expression was also correlated with immune cell infiltration and caused poor outcomes depending on the abundance of tumor-infiltrating immune cells in ccRCC. Our findings suggest that cyclin F/CCNF expression is likely to have an essential role in ccRCC pathobiology through regulating multiple oncogenic signaling pathways and affecting the tumor immune microenvironment and may serve as prognostic biomarker and promising therapeutic target in ccRCC.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) accounts for approximately 2% of cancer-related diagnoses and deaths globally. Its incidence rate has increased in the last decades, attributed to incidental diagnoses associated with the extensive use of imaging techniques1,2. Clear-cell RCC (ccRCC) represents the predominant and simultaneously the most lethal histological subtype of RCC tumors3. When diagnosed early, ccRCC can be successfully managed with surgery. Nevertheless, due to its typically asymptomatic nature, nearly 28% of cases are initially diagnosed with regional or distant metastases4.
Moreover, up to 30% of the patients with initially localized disease experience recurrence after cancer resection5,6, and the high heterogeneity of ccRCC as well as its resistance to radiotherapy and chemotherapy, renders available systemic therapies largely ineffective7. Consequently, 5-year overall survival in the IV stage of ccRCC is 19.7%8. In the face of difficulties in the early detection of ccRCC and the inadequate effectiveness of non-surgical methods, identifying new reliable diagnostic and prognostic biomarkers seems crucial for improving patient outcomes.
Cyclin F, also known as F-box only protein 1 (Fbxo1), unlike the other cyclins, does not act as an activator of cyclin-dependent kinases (CDKs). Instead, it is a member of the F-box proteins being substrate recognition subunits of SCF (Skp1-Cul1-F-box protein) ubiquitin ligase complexes. Consequently, cyclin F is implicated in various target proteins’ ubiquitination and subsequent degradation. The protein level of cyclin F increases in the S phase and peaks in the G2 phase of the cell cycle, which is strictly regulated to ensure the scheduled degradation of substrates9. The altered expression of cyclin F has been shown to be associated with poor prognosis of patients with various types of cancers10,11,12,13,14 as well as the growth, metastasis, and drug resistance in some of them. Considering these findings, cyclin F emerges as a potential prognostic marker and therapeutic target. However, its clinical significance in ccRCC remains unexplored.
In this paper, we explore the clinical association of cyclin F/CCNF with ccRCC pathology and outcome using two independent tissue array ccRCC cohorts and TCGA ccRCC cohort, followed by further validation of the findings with additional bioinformatics analyses. In addition, the mechanisms underlying how CCNF affects ccRCC were examined by analyzing the correlations between CCNF and other genes followed by functional enrichment analysis.
Materials and methods
Ethics statement
The usage of archived diagnostic leftover tissues for research purposes, as well as patient data analysis, was approved by the Bioethics Committee of the Nicolaus Copernicus University in Toruń functioning at Collegium Medicum in Bydgoszcz (no. 253/2018). The need for informed consent was waived by the same Bioethics Committee as mentioned above (Bioethics Committee of the Nicolaus Copernicus University in Toruń functioning at Collegium Medicum in Bydgoszcz) in view of the retrospective nature of the study and all procedures performed were part of routine care, and the study utilized only leftover tissues, with the analysis using anonymous clinical data. Data collection and management were performed in compliance with the principles of the Declaration of Helsinki.
Tissue arrays and immunochemistry staining
The study was conducted retrospectively in two independent tissue arrays (TAs) cohorts of ccRCC patients. The first one consisted of 108 ccRCC cases and 37 adjacent non-tumor tissues obtained from the patients who underwent surgery in the Department of Urology and Andrology, Antoni Jurasz University Hospital No. 1 in Bydgoszcz (Poland) and were subsequently diagnosed with ccRCC in the Department of Clinical Pathomorphology, Collegium Medicum in Bydgoszcz of Nicolaus Copernicus University in Torun; whereas the other one was commercially purchased (tissue microarray (TMA) no. KD20810a, TissueArray.Com LLC, Derwood, MD, USA) and contained 192 ccRCC tumors and 16 normal kidney samples (1 mm diameter spot per patient). Tissue macroarrays (TMAs) for the former cohort were constructed according to the previously described method10. Each paraffin recipient block containing five different large (about 5–7 µm) tissue fragments was sectioned at 4-μm thickness and placed on high-adhesive glass slides (SuperFrost Plus; Menzel-Glaser, Braunschweig, Germany). The institutional tissue macroarray cohort was thereafter referred to as the TMA_1 cohort, while the commercial microarray cohort was called TMA_2. Tissue sections of both cohorts were stained by the same previously described procedure10. They were incubated with a primary rabbit polyclonal anti-cyclin F antibody (1:100, 40 min; cat. no: HPA071600, Merck Millipore, Burlington, MA, USA) on BenchMark ULTRA system (Roche Diagnostics/Ventana Medical Systems, Tucson, AZ, USA). Antigen–antibody reactions were visualized using an ultraView Universal DAB Detection Kit (Roche Diagnostics/Ventana Medical Systems, Tucson, AZ, USA). The complete clinicopathological characteristics of experimental groups are presented in Supplementary Table S1 online. Overall survival (OS) was defined as the time interval between the date of surgery and date of death. OS data were censored if patients were alive at the last follow-up date (21 January 2021). The median follow-up time (calculated by the reverse Kaplan–Meier method) and median OS time for patients in the TMA_1 cohort were 181 and 42 months, respectively.
Evaluation of immunostaining
TMA sections were examined for immunoreactivity and graded semi-quantitatively using the H-score system by three investigators in a blinded fashion under a multi-headed microscope (Olympus, Tokyo, Japan) at 20 × original objective magnification. The H-score was calculated by adding the products of the percentage of cells stained at a given staining intensity (0–100) and the staining intensity score (1, weak, 2, moderate, and 3, strong), resulting in a range from 0 to 300. The H-score was calculated as follows: ∑ (pi × i) = (1 × percentage of weak intensity) + (2 × percentage of moderate intensity) + (3 × percentage of strong intensity). To determine "high" or "low" expression levels of cyclin F in the TMA_1 cohort, the final H-scores were dichotomized based on the optimal cut-off point, as well as the median (H-score ≤ 21.5, low expression; H-score > 21.5, high expression; data provided in Supplementary files). The optimal cut-off value for cyclin F expression was determined using the cutp algorithm in the Evaluate Cupoints software15 and classified as follows: H-score < 11, low expression; H-score ≥ 11, high expression. Patients of the TMA_2 cohort were divided into a low cyclin F expression group (H-score ≤ 5) and high cyclin F expression group (H-score > 5) based on the median value of the H-score.
Extraction of RNA-sequencing TCGA data and clinical information
The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases provided data on CCNF expression for 475 ccRCC primary tumors and 28 normal tissue samples, respectively. The baseline characteristics of these patients are presented in Supplementary Table S1. The above data, normalized using the DESeq2 method, were downloaded through the University of California Santa Cruz (UCSC) Xena Browser (https://xenabrowser.net/, accessed on 14 September 2022)16, whereas corresponding clinical information was retrieved from cBioPortal (https://www.cbioportal.org, accessed on 14 September 2022). According to the optimal cutpoint determined for CCNF expression levels using the cutp function of the Evaluate Cutpoints software15, ccRCC patients were divided into high (≥ 7.884) and low (< 7.884) expression groups. The median OS time for patients in the TCGA cohort was 116 months.
TNMplot analysis
The TNMplot database (http://tnmplot.com; accessed on 28 November 2022) was applied to compare the CCNF gene expression in paired tumor and adjacent normal tissues, as well as between normal, tumor, and metastatic tissues. This analysis utilized the gene chip data obtained from the National Center for Biotechnology Information Gene Expression Omnibus Database (NCBI-GEO)17. Besides, CCNF expression levels were also compared in TCGA tumor samples paired with adjacent TCGA normal samples based on RNA-seq data. The results were visualized by violin plots. CCNF expression in normal, tumor, and metastatic tissues was compared using Kruskal–Wallis test with Dunn's post hoc tests, whereas the difference between two groups (tumor vs. control tissue) was analyzed with a Mann–Whitney U test or a paired Wilcoxon statistical test.
ShinyGEO
The ShinyGEO (http://gdancik.github.io/shinyGEO/, accessed on 30 November 2022) was used for further differential gene expression analysis, where GSE15641, GSE16449, GSE66272, GSE1963, and GSE105261 were employed18. Boxplots were used to depict the distribution of gene expression levels. The Mann–Whitney or Wilcoxon test or Kruskal–Wallis test with Dunn's post hoc test was performed to assess whether the expression difference was statistically significant (p < 0.05).
UALCAN web portal
The "TCGA" module of the UALCAN online tool (http://ualcan.path.uab.edu/, accessed on 30 November 2022) was used to assess the relative expression of CCNF in different grades of kidney renal clear cell carcinoma (KIRC)19. The results were presented in box-whisker plots, and differences in transcriptional expression were compared by Student's t-test considering unequal variance. P < 0.05 was considered statistically significant.
GENT2, GEPIA2, Kaplan plotter online databases
GENT2 and GEPIA2 online tools were used to validate the prognostic value of CCNF expression in RCC cohorts derived from the NCBI-GEO public database and TCGA, respectively. Meta-survival analysis of OS with Cox proportional hazard modes was done using the data from the Gene Expression across Normal and Tumor tissue (GENT2) (http://gent2.appex.kr/gent2/; accessed on 29 November 2022) and depicted as a forest plot20. The Gene Expression Profiling Interactive Analysis (GEPIA2, http://gepia2.cancer-pku.cn/, accessed on 13 December 2022) was used to obtain the OS plots of the candidate gene across KIRC-TCGA tumors21. Median and quartile values [cutoff-high (50%, 75%, respectively) and cutoff-low (50%, 25%, respectively)] were set as the expression thresholds for splitting the high-expression and low-expression cohorts. Survival curves with the calculated HRs with 95% CIs and the log-rank p-values were plotted. GEPIA2-based analysis was also used to determine the correlation between CCNF expression and immune infiltration associated markers (accessed on 8 December 2023). The correlation was analyzed by Spearman's correlation coefficient. In addition, the online Kaplan–Meier Plotter tool (http://kmplot.com/analysis, accessed on 13 December 2022), which combines survival data from GEO, TCGA, and other databases, was applied further to confirm the effect of CCNF on ccRCC prognosis22. Kaplan–Meier curves were plotted using the "mRNA RNA-seq" module and "CCNF" as input queries; patients were divided based on the best cut-off and median. Analyses were or were not filtered by tumor stage. A statistically significant difference was considered when a p-value < 0.05.
TIMER2 database analysis
In the present study, we conducted the Tumor IMmune Estimation Resource 2.0 (TIMER2; accessed on 8 December 2023)23 to confirm the expression and prognostic value of CCNF in TCGA-KIRC. Moreover, we applied TIMER2 to determine the correlation between CCNF expression and the abundance of tumor-infiltrating immune; however, we focused only on those immune cells that were significantly associated with the overall survival via the ‘Outcome_Module’ of TIMER2. The ‘Outcome_Module’ was also used to explore the difference in overall survival among patients stratified by both the estimated infiltration level of immune cells and CCNF expression level in KIRC. Given that the tumor purity of the sample impacts the immuno-infiltration analysis, the correlation analysis was adjusted accordingly. All analyses were performed with default parameters in TIMER2 and p-value < 0.05 was considered statistically significant.
PPI construction and functional enrichment analysis
The list of genes positively and negatively correlated with CCNF in ccRCC was retrieved from the UALCAN database (http://ualcan.path.uab.edu/, accessed on 23 March 2023)19. The top positively correlated genes (Pearson CC values ≥ 0.7) and all negatively correlated genes (Pearson CC values ≥ 0.3) were selected to construct the zero-order protein–protein interaction network (PPI) using The Search Tool for the Retrieval of Interacting Genes Database (STRING)24 and Cytoscape software (version 3.9.1). The top 10 genes in the network were screened as hub genes using three topological analysis methods of the CytoHubba plugin (version 0.1) in the Cytoscape software25. The Molecular Complex Detection (MCODE) clustering algorithm (version 2.0.2) was then used to identify clustered modules within the PPI network according to the following criteria: degree cut-off = 2, max. depth = 100, k-core = 2, haircut = yes, and node score cutoff = 0.226. The most significant subnetwork was tested for overrepresentation of the Gene Ontology (GO) terms using the DAVID Functional Annotation Clustering Tool (The Database for Annotation, Visualization and Integrated Discovery version 6.8, DAVID; https://david.ncifcrf.gov, accessed on 23 March 2023)27, whereas the ClueGO plugin (version 2.5.9) was used to identify enriched KEGG and Reactome pathways (with a threshold of p ≤ 0.0001 based on a two-sided hypergeometric test (kappa score 0.4) and the Bonferroni correction)28.
Statistical analysis
All statistical analyses were conducted using GraphPad Prism (version 8.0, GraphPad Software, San Diego, CA, USA), SPSS (version 28.0, IBM Corporation, Armonk, NY, USA), or Rstudio (version 1.3.1093) software packages. Data distribution was determined with the Shapiro–Wilk test, and the appropriate parametric or nonparametric statistical tests were applied. The Mann–Whitney or Wilcoxon signed-rank test was performed to compare differences in cyclin F expression between experimental and control groups. The chi-square or Fisher's exact test was employed to evaluate differences between cyclin F expression status and clinicopathological parameters. The Kaplan–Meier curves and log-rank test were utilized to compare survival outcomes in different groups of ccRCC patients. The prognostic ROC curves were constructed from Kaplan–Meier survival estimates, following the method described by Combescure et al.29. Univariable and multivariable analyses by Cox proportional hazards method were performed to establish the risk factors for the overall survival. Crude and adjusted hazard ratios (HRs) with a relative 95% confidence interval were shown. A backward elimination procedure built all multivariable models, with a significance level of p < 0.05 to enter the model and p < 0.10 to stay. The proportional hazard assumptions were verified graphically using plots of log(−log) survival and Schoenfeld residuals. Covariates violating the PH assumption were introduced as time-dependent covariates in the Cox regression models. All reported p-values were two-sided with a significance level of 0.05.
Results
Expression levels of cyclin F/CCNF in ccRCC and control tissues
We first determined the IHC expression of cyclin F in two independent TMA cohorts: in-house cohort (TMA_1), consisting of 108 ccRCC tissues and 37 adjacent non-tumor tissues, and the other, commercially purchased one (TMA_2), encompassing 192 ccRCC cases and 16 samples of normal renal tissue. After excluding non-representative and missing cores, IHC results were available for 108 (100%) and 172 (89.58%) cases of TMA_1 and TMA_2 cohorts, respectively.
Faint cytoplasmic staining of cyclin F was occasionally observed in a renal tubular epithelium of adjacent non-tumor tissues but not in a renal tubular epithelium of normal tissues (Supplementary Fig. S1). In control tissues, mild to moderate cyclin F labeling was noticed in endothelial cells of glomerular capillaries and, to a lesser frequency, peritubular capillaries, as opposed to the much more common and varied in intensity (often strong) staining of endothelial cells of tumor-associated vasculature (Supplementary Fig. S1). In the tumor tissues of both cohorts, cyclin F staining was predominantly cytoplasmic, with mixed membranous-cytoplasmic staining or solely membranous staining observed in the same cases (Supplementary Fig. S1). In addition, we identified noticeable inter- and intratumoral differences in IHC staining intensity, distribution, and patterns, which could be partially due to the glycogen and lipid-rich cytoplasmic deposits being washed away during tissue processing. Statistical analysis showed a significant increase of cyclin F expression in ccRCC compared to tumor-adjacent normal tissue (Fig. 1a) or normal renal tissue (Fig. 1b). According to the established cut-off points, high expression levels of cyclin F were found in 74 (68.52%) and 78 (45.35%) cases of ccRCC in the TMA_1 and TMA_2 cohorts, respectively.
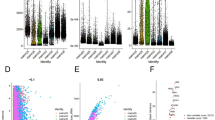
Expression of cyclin F/CCNF in clear cell renal cell carcinoma compared to non-cancerous control tissue. Boxplot graphs of (a) cyclin F expression in tumor tissue (n = 108) and histologically normal tissue adjacent to the tumor (n = 37) of the institutional tissue macroarray (TMA) cohort based on immunohistochemistry (IHC); (b) cyclin F expression in tumor tissue (n = 172) and normal renal tissue (n = 16) of the commercially purchased microarray (TMA) cohort based on IHC; (c) CCNF expression levels in tumor tissue (n = 475) and normal renal tissue (n = 27) based on RNA-Seq data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, respectively, downloaded from UCSC Xena Browser. Violin plots from the TNMplot web tool showing (d) CCNF expression levels in paired tumor tissue (n = 72) and adjacent normal tissue (n = 72) based on RNA-Seq data from the TCGA; (e) CCNF expression levels in paired tumor tissue (n = 210) and adjacent normal tissue (n = 210) based on the gene chip data derived from the Gene Expression Omnibus (GEO) database. (f) CCNF expression levels in primary tumor tissue (n = 556), non-paired normal tissue (n = 277), and metastasis (n = 58) based on the gene chip data derived from the GEO database.
Consistent with the protein data, the RNA-Seq data from the TCGA showed that the CCNF expression was significantly increased in ccRCC tissues relative to normal renal tissues (Fig. 1c). Similar results were obtained from the TNMplot online tool based on the TCGA-RNA-seq data when comparing paired ccRCC and histologically normal tissue adjacent to the tumor (Fig. 1d). Likewise, CCNF expression was higher in tumor tissues than in matched adjacent normal tissues (Fig. 1e) or non-paired normal tissue (Fig. 1f) but not in metastasis (Fig. 1f) from the gene chip data of the TNMplot. Notably, CCNF expression in metastases was significantly lower by a factor of 0.85 relative to primary tumors (Fig. 1f). However, a differential expression analysis of individual GEO datasets through the ShinyGEO web-based application found that CCNF expression was increased when compared to control and tumor tissues and between primary tumor and metastatic tissues; nevertheless, in the latter case, the statistical significance was not reached (p = 0.134; Supplementary Fig. S2).
Association with clinicopathological features
In the TMA_1 cohort (Table 1), ccRCCs with increasing tumor grade more frequently had cyclin F-high than cyclin F-low expression (p = 0.018). Furthermore, high-level cyclin F expression was marginally significantly linked to more advanced pT stages (p = 0.055). No other associations were found between cyclin F and investigated clinicopathological parameters.
In the TMA_2 cohort (Table 1), ccRCC patients with histologic grade 2 and 3 tumors more frequently had cyclin F-high expression (69.23%) than low expression (30.77%; p = 0.017). Furthermore, pT2-pT4 tumors tended to be cyclin F high expressors (56.36%) than cyclin F low expressors (43.64%), while in pT1 tumors, it was the opposite (40.17% vs. 59.83%; p = 0.051). A similar association was found regarding the TNM stage (p = 0.034). No other relationships were observed between cyclin F and clinicopathological features.
In the TCGA cohort (Table 2), ccRCC patients affected by advanced T and TNM stage (both p < 0.0001), or those with a proven positive lymph node (p = 0.014), or M1 disease (p < 0.0001) more frequently had tumors with CCNF-high expression than CCNF-low expression. Moreover, there was a trend toward the association of CCNF status with tumor grade (p = 0.107). Notably, when analyzed as a continuous variable using the UALCAN web tool, CCNF expression increased with increasing tumor grade (Supplementary Fig. S3). Statistical significance was reached between the subgroups, except for grade 1 vs. grade 2 tumors (Supplementary Fig. S3). No other significant associations existed between the tested biomarker and remaining clinicopathological features, including age and sex.
Association with the clinical outcome
In Kaplan–Meier analysis of the TMA_1 cohort, cyclin F-high expression was significantly associated with shortened median OS (34 months) compared with cyclin F-low (77 months; p = 0.003; Fig. 2a). As presented in Table 3, statistically significant univariable hazard ratios (HRs) were found for cyclin F (HR 1.91, 95% CI 1.23–2.98), age at diagnosis (HR 1.63, 95% CI 1.09–2.46), sex (HR 0.57, 95% CI 0.37–0.88), tumor grade (HR 3.46, 95% CI 1.90–6.31), and nodal status (HR 3.44, 95% CI 1.55–7.64). In a subsequent stepwise selection analysis, all these remained independent prognostic factors for OS (Table 3; all p < 0.05). The area under the prognostic ROC curve was 0.66: cyclin F overexpressors had a 66% probability of dying before cyclin F low expressors (Fig. 2b).
Effect of cyclin F expression on survivals in clear cell renal cell carcinoma cohort. Cases were divided into low and high-expression groups according to the optimal cut-off point (Cp = 11) determined by the Evaluate Cutpoints software. (a) Kaplan–Meier curve comparing high and low expression of cyclin F in the entire TMA_1 cohort (n = 108) and the corresponding (b) prognostic ROC curve. The area under the prognostic ROC curve (AUC) equals the probability that the time-to-event is shorter in high-risk patients than in low-risk patients29. The number of cases (n) and events (e) for low and high-expression groups is displayed. The p-value was calculated by the log-rank test.
Univariable (Supplementary Fig. S4) and multivariable (Supplementary Table S2) comparisons of the H-score at the median cut-off showed similar associations with the OS.
In Kaplan–Meier survival analysis of the TCGA cohort, CCNF-high expression was significantly associated with a shorter OS in the whole cohort (52 months vs. not reached; p < 0.0001; Fig. 3a) as well as across all TNM stages (all p < 0.01; Fig. 3c–e). The areas under the prognostic ROC curve were 0.785 (an optimistic scenario) and 0.744 (a noninformative scenario) (Fig. 3b). In unadjusted Cox proportional hazards regression (Table 4), CCNF (HR 3.52, 95% CI 2.50–4.96), TNM stage (HR 3.61, 95% CI 2.59–5.02), T classification (HR 3.19, 95% CI 2.31–4.39), N classification (HR 3.78, 95% CI 2.01–7.13), and metastasis (HR 4.23, 95% CI 3.05–5.86) were significantly associated with OS. Importantly, multivariable Cox analysis demonstrated that CCNF remained a highly significant predictor of OS independently of histologic grade and TNM stage (HR 2.86, 95% CI 2.02–4.06, p < 0.0001; Table 4). Adjustment for pT, pN, and pM categories instead of AJCC disease stages yielded similar results for the CCNF expression (Supplementary Table S3). Further analysis through bioinformatics web resources, including GEPIA2, Kaplan–Meier Plotter, and GENT2 (Supplementary Fig. S5), confirmed that CCNF expression was highly discriminant for OS of ccRCC patients.
Effect of CCNF expression on survivals in The Cancer Genome Atlas (TCGA) data portal cohort. Cases were divided into low and high-expression groups according to the optimal cut-off point (Cp = 7.884) determined by the Evaluate Cutpoints software. (a) Kaplan–Meier curve comparing high and low expression of CCNF in the entire TCGA cohort (n = 475) and the corresponding (b) prognostic ROC curve. The area under the prognostic ROC curve (AUC) equals the probability that the time-to-event is shorter in high-risk patients than in low-risk patients. The missing part of the prognostic ROC curve (the straight dashed line) was extrapolated with optimistic (AUC1) and noninformative (AUC2) assumptions29. Kaplan–Meier curves comparing high and low expression of CCNF in (c) stage I subgroup (n = 234), (d) stage II–III subgroup (n = 169), and (e) stage IV subgroup (n = 72). p-values were calculated by the log-rank test. The number of cases (n) and events (e) in low and high expression groups is displayed.
Association with tumor immune infiltration
Here, we investigated the potential of CCNF to display the prognostic significance of immune cells in KIRC. Using ‘Gene_DE’ and ‘Gene_Outcome’ modules of the TIMER2 web server, we first confirmed that CCNF was significantly upregulated in KIRC relative to adjacent normal kidney tissue and that CCNF-high expression reflected a poor clinical outcome in KIRC (Supplementary Fig. S6). Then, we demonstrated that CCNF expression had significant positive correlations with infiltrating levels of activated natural killer (NK) cells, T cell NK cells, follicular helper T (Tfh) cells, regulatory T (Tregs) cells, and myeloid-derived suppressor cells (MDSCs); and the patients with higher infiltration of these immune cells indicated an unfavorable prognosis (Supplementary Fig. S7). In turn, CCNF was negatively correlated with resting memory CD4 + T cells, activated mast cells, and hematopoietic stem cells (HSCs); and the patients with lower infiltration of these immune cells had an adverse prognosis (Supplementary Fig. S7). In addition, CCNF expression demonstrated a significant negative correlation with tumor purity in KIRC (r = − 0.122, p = 0.00853; data not shown). Importantly, KIRC patients with high CCNF expression and high NK cell infiltration had significantly worse prognosis than those with high CCNF expression and low infiltration of NK cells (HR = 1.58, p = 0.0164; Fig. 4a). For patients with low CCNF expression, high NK cell infiltration indicated poorer survival than those with lower infiltration level (HR = 1.6, p = 0.0335; Fig. 4a). Furthermore, KIRC patients with a particularly poor prognosis had both high expression of CCNF and increased infiltration degree of T cell NK cells (Fig. 4b), Tfh cells (Fig. 4c), Tregs (Fig. 4d), or MDSCs (Fig. 4e). Likewise, tumors with concomitant high expression of CCNF and lower infiltration of resting memory CD4 + T cells (Fig. 4f), activated mast cells (Fig. 4g), or HSCs (Fig. 4h) also experienced the worst prognosis.
The association between survival and clear cell renal cell carcinoma defined by CCNF expression and immune infiltration level. Kaplan–Meier plots from the ‘Outcome Module’ of the Tumor IMmune Estimation Resource 2.0 (TIMER2) show the difference of overall survival among patients stratified by both CCNF expression level the estimated infiltration level of (a) activated NK cell; (b) T cell NK; (c) follicular helper T cells; (d) regulatory T (Tregs) cells; (e) myeloid-derived suppressor cells (MDSCs); (f) resting memory CD4 + T cells; (g) activated mast cells, and (h) hematopoietic stem cells (HSCs). The immune cell infiltration and CCNF expression were divided into high and low groups according to the median. The hazard ratio and the log-rank p value are shown.
Moreover, GEPIA2 was applied to explore the correlation between CCNF expression and gene markers of tumor-infiltrating immune cells in TCGA-KIRC. In particular, Treg-related gene signature had the strongest correlations with CCNF expression (r = 0.47, p < 0.0001; Supplementary Fig. S8). The results of GEPIA2 additionally showed the positive correlations between CCNF and TAM-associated markers, as well as M1 and M2 macrophage markers (Supplementary Fig. S8). We also determined the relationship between CCNF expression and T cell exhaustion-associated markers, including HAVCR2 (TIM-3), TIGIT, LAG3, PD-1 (PDCD1), CXCL13, LAYN, CTLA4, GZMB. The results demonstrated that the CCNF expression level was positively correlated with the T-cell exhaustion signature in TCGA-KIRC (Supplementary Fig. S8).
PPI network construction and functional enrichment analysis
Ninety-one genes positively correlated with CCNF and 30 genes negatively correlated with CCNF met the inclusion criteria. The PPI network consisted of a total of 100 nodes with 2917 edges (clustering coefficient 0.865, PPI enrichment p-value: < 1.0e−16), including 91 nodes positively correlated with CCNF and nine nodes negatively correlated with CCNF (Fig. 5a); the remaining nodes represented negatively correlated genes, which evidently were disconnected, as shown in Fig. 5b. The top 10 nodes with the highest degree, betweenness centrality, and closeness centrality are listed in Fig. 5c. The two clusters identified by MCODE are listed in Fig. 5d; this with a high clustering score (68.535) is shown in Fig. 5e. Functional enrichment analysis was then applied to the genes of cluster 1 to gain insights into the role they may play in ccRCC. The most significant (FDR-corrected p-value < 0.01) "Biological Process", "Cellular Components", and "Molecular functions" GO terms are listed in Supplementary Table S4. The enriched Reactome Pathways and KEGG terms related to the queried genes are depicted in Fig. 5f.
Functional enrichment analysis for CCNF-correlated genes. (a) Protein–protein interaction (PPI) network for the top genes positively (green nodes) or negatively (blue nodes) correlated with CCNF in clear cell renal cell carcinoma; (b) disconnected nodes; (c) The top 10 hub genes identified by CytoHubba Cytoscape plugin are ranked according to degree centrality, betweenness centrality, and closeness centrality; (d) The two modules identified in the PPI network using the MCODE Cytoscape plugin. (e) The top module (cluster 1) identified from the PPI network using the MCODE Cytoscape plugin; (f) Enrichment of nodes in the PPI network by Reactome Pathways and KEGG terms is illustrated using the ClueGO plugin.
Discussion
Cyclins, as master cell cycle regulators, and since their discovery in 1982, are in the spotlight of cancer research. Initially, cyclins were thought to be simple switches enabling cell cycle progression. Still, years of research have shown that their function is much more complex and can affect many cellular processes. For example, recent studies show the involvement of the cyclins and Cdks in molecular pathways associated with the regulation of transcription, epigenetic regulation, metabolism, stem cell self-renewal, neuronal functions, and spermatogenesis30. Increasing evidence also suggests the involvement of cyclin F/CCNF in cancer. Nevertheless, the data indicate that cyclin F is a multifaceted protein with an ambiguous role in cancer cells. In this study, we investigated the prognostic value of cyclin F/CCNF in ccRCC patients.
As far as we know, prior to our study, the prognostic value of cyclin F protein had not yet been investigated in the context of ccRCC. However, CCNF gene expression has recently been reported as a promising biomarker for prognosis prediction in ccRCC31,32,33,34. Here, by integrating analyses through IHC, RNA-seq, and gene chip microarray data, we reproducibly found the remarkable differences in CCNF protein and gene expression levels between tumor and control tissues. Furthermore, cyclin F immunoreactivity was observed in most ccRCCs, in a minority of tumor-adjacent normal tissues, and neither of normal renal tissues. Moreover, tumors with features of more aggressive behavior were more likely to display cyclin F-high expression than low. These results suggest that cyclin F overexpression may provide a selective advantage in ccRCC pathobiology. Furthermore, they further support the notion that normal adjacent to tumor tissue represents a unique intermediate state between healthy and tumor tissue35. Finally, they also highlight the attractiveness of cyclin F as a biomarker in ccRCC, as the on–off expression pattern facilitates the IHC staining interpretation in routine practice36.
Previous studies have shown that altered CCNF expression is associated with a poor prognosis in various cancers, including breast, liver, pancreatic, brain, and skin cancers12,13,14, 37. In the current study, the analysis of the data from the TCGA database reveals that 45.89% of ccRCC patients are characterized by high CCNF mRNA expression. Furthermore, overexpression of CCNF in these individuals is associated with a poorer prognosis, advanced T and TNM stage, and distant as well as lymph node metastasis. Moreover, the expression of CCNF increases proportionally with tumor grade. Indeed, Wei et al. have recently shown that the mRNA expression levels of CCNF in KIRC-TCGA increased gradually with a disease progression and correlated with adverse OS and RFS31. These observations are also in agreement with some other studies investigating CCNF mRNA levels in different tumors. Our former analysis on melanoma and pancreatic adenocarcinoma also revealed that high CCNF mRNA level was associated with a worse prognosis for the patients12,37. Similarly, the study presented by Liu and colleagues demonstrated a strong relationship between high CCNF mRNA levels and worse prognosis in breast cancer (BC) patients. Moreover, CCNF overexpression significantly correlated with worse RFS, OS, and DMFS among all types38. Importantly, these observations have been confirmed on the protein level where high expression of cyclin F was observed in Luminal A, Luminal B, HER2, and TNBC breast cancer. Additionally, the in vitro experiments showed that silencing cyclin F reduces proliferation rate and migration of breast cancer cells38. Another example is liver cancer, where an elevated level of CCNF was associated with worse OS. Additionally, this overexpression cannot be led by the mutations in the CCNF gene because only 0.81% of 372 liver cancer patients bear the mutation in CCNF locus14.
Interestingly, the IHC study by Fu et al. on in-house liver cancer series showed the opposite, including the association of cyclin F-low with more aggressive tumor features and a prognosis11. The liver cancer studies mentioned above11,14 present the opposite prognostic value of cyclin F depending on the source of data or, more precisely, the measurement level (protein vs. mRNA), leading to further controversy regarding the functional portrait of this cyclin in human cancers. We witnessed similar discrepancies in our studies on melanoma and pancreatic adenocarcinoma. The bioinformatics analysis of melanoma patients pointed to CCNF as a negative prognostic marker, and it was reasonable to consider CCNF as an oncogene. However, the in vitro studies do not confirm the assumption that CCNF is an oncogene. After cyclin F silencing, we observed an increased proliferation rate and elevated capabilities for migration and invasion. Additionally, the morphological and molecular alterations suggested the induction of the process similar to epithelial-to-mesenchymal transition in the A375 melanoma cell line12,39. In pancreatic adenocarcinoma, high CCNF mRNA was associated with poor prognosis, whereas the univariable Cox proportional hazards model pointed to cyclin F protein as a favorable prognostic marker; however, the last observation was not statistically significant37. Here, we obtained consistent results from both TCGA and TMA cohorts. TCGA and TMA_1 cohorts are characterized by reduced OS in cyclin F-high groups. Furthermore, in TMA and TCGA cohorts, patients with more aggressive tumor features more frequently had cyclin F/CCNF-high than cyclin F/CCNF-low expression. All presented data suggest a potential oncogenic role for cyclin F in clear cell renal cell cancer . By contrast, Deshmukh et al. presented the oncosuppressive properties of cyclin F in glioma xenografts. The authors observed a decrease in cyclin F expression simultaneously with the increase in tumor grade40.
To shed more light on the underlying mechanisms behind the prognostic value of CCNF expression in ccRCC, we identified CCNF-correlated genes, and then built PPI network and performed functional enrichment analyses of the most significant gene cluster. All of the nodes of cluster 1 genes are highly interacting and therefore act as hub (high degree) genes in the PPI network, and may be corresponding to the ccRCC-causing genes41. As can be seen, the vast majority of the hub genes identified in the PPI by calculating centrality parameters (using the CytoHubba plugin) are also present in the cluster 1, which futher emphasizes their role in cancer-relevant pathways. Generally, cluster 1 showed the enrichment for gene sets associated with cell cycle and cell division, in particular M phase and mitotic checkpoint, being highly relevant to the control of cell growth and proliferation. In this context, it is essential that overexpression of CCNF is positively correlated with proliferation-promoting genes, suggesting that it contributes to cancer progression and therefore poor patient prognosis by promoting excessive cell proliferation. Indeed, many of our hub genes are already known from their ocogenic and pro-proliferative activities in ccRCC. The node TTK possesses the highest degree centrality and has been shown to play an important role in maintaining the proliferative and invasive potential of ccRCC cells, both in vitro and in vivo in mice42. In our PPI network, TTK interacts with UBE2C43, CDCA844, KIF2C45, PLK146, AURKB47, NCAPG48, CCNB149, CDK148, NDC8050, all of which have been previously implicated in the initiation and/or progression of ccRCC. Furthermore, many of the upstream or downstream regulators of the listed genes may activate deleterious signaling (e.g. phosphorylation) pathways, having established connections with human cancers, including ccRCC. Also notably, these highly-related genes in the PPI network may reciprocally control each other or be affected by the same transcription factors, thus targeting one may interrupt the entire cluster and even the whole PPI network. For example, UBE2C, which is detected as the second pivotal hub gene in the present study, is a transcriptional target of FOXM1, which is also included in cluster 1. On the other hand, transcriptional activity of FOXM1 is strongly dependent on phosphorylation by PLK, which therefore controls the execution of the transcriptional program required for mitotic progression of ccRCC cells46. FOXM1 is proven to be one of the key transcription factors regulating oncogenic signalling pathways during the ccRCC progression. Together with UBE2C, it activates AKT/mTOR signaling pathway to increase the cell cycle progression, proliferation rate, cell survival, as well as the migratory and invasive abilities of ccRCC cells51. In addition, FOXM1/KIF20A axis executes oncogenic activity in ccRCC via activation of the EMT signaling52, while FOXM1/AURKB axis via stimulation of cancer cell proliferation51. Importanlty, both KIF20A and AURKB are included in cluster 1 genes. AURKB decreases the expression of the cell cycle inhibitor P21 by inhibiting P53 activity, subsequently causing upregulation of CDK1, eventually leading to cell division and increasing the tumor cell survival53. CDK1 is also affected by the proportion of FOXM1 and KIF20A overexpression54, which further highlights their decisive roles in the cell division and mitosis. In fact, our functional analyses are consistent with recent in silico and in vitro studies of Li et al., who showed that CCNF might promote ccRCC proliferation by inhibiting cell senescence through the CDK1-P53 signaling pathway33. Taken together, it appears that CCNF and its highly correlated genes initiate the signaling pathways that eventually result in uncontrolled cell proliferation, thus targeting the cyclin F offers a potential novel strategy to target the ccRCC cells efficiently.
Another aspect of this study was to estimate the relevance between the CCNF and tumor immune infiltration in TCGA-KIRC. In agreement with the recent studies of Gao et al.32, as well as Wei et al.31, our analysis showed that CCNF was closely associated with immune cell infiltration in TCGA-KIRC. Interestingly, CCNF was positively correlated with the infiltration abundance of activated NK cells, TNK cells, Tfh cells, Tregs, and MDSCs, the high expression of which predicted poor survival in KIRC-TCGA patients, while it was negatively correlated with resting memory CD4 + T cells, activated mast cells, and HSCs that conferred a better prognosis when present in tumor microenvironment. Furthermore, here we also explored the difference in overall survival among KIRC patients stratified by both the estimated infiltration level of immune cells and CCNF expression level. Specifically, we found that KIRC patients with a particularly poor prognosis had concomitantly high expression of CCNF and increased infiltration of NK cells, TNK cells, Tfh cells, Tregs, MDSCs, or decreased abundance of resting memory CD4 + T cells, activated mast cells, or HSCs. These results suggest that immune cell infiltration may be one of the reasons that caused CCNF to become a prognostic factor in ccRCC. Furthermore, in addition to having the strongest correlations with Treg-related gene signature, CCNF expression was also highly significantly (p < 0.0001) associated with gene markers of, for example, the M2 macrophages and exhausted T cells. We can therefore speculate that CCNF is involved in immune escape and immunosuppression in the ccRCC microenvironment, and it may serve as a potential target to increase the effectiveness of immunotherapy in ccRCC. However, the abundance and gene markers of different tumor-infiltrating immune cells were found to be only weakly to moderately correlated with CCNF (the range for correlation coefficients: 0.1–0.47). This is probably why, we did not find CCNF-correlated genes to be highly enriched in immune processes and immune-related pathways via our functional enrichment analysis, which was set at high confidence.
Our study has some limitations that weaken the conclusiveness of the presented data. First, it is impossible to directly correlate the protein data from our cohort and the RNA-seq data from the TCGA cohort. Simultaneously analyzing mRNA and protein from the same sample would significantly improve the quality of the conclusions by providing a more comprehensive understanding of the relationship between mRNA expression and protein levels. Also, the structure of the cohorts differs from each other. For example, in the TMA_1 cohort, 65.74% of the patients were diagnosed with grade 2 ccRCC. On the other hand, in TMA_2, 80.23% were diagnosed with grade 1 ccRCC, whereas in the TCGA cohort, 54.95% were diagnosed with grades 3–4. Moreover, although the present study goes some way towards understanding the potential mechanisms underlying how CCNF affects ccRCC, it is clear that our bioinformatics analyses still need further verification through in vivo and in vitro experiments. Notwithstanding the described limitations, our study provides strong evidence for the utility of cyclin F as a prognostic and diagnostic marker in ccRCC.
In conclusion, by analyzing protein levels and publicly available transcriptomic data, we found that cyclin F/CCNF is significantly upregulated in ccRCC patients. High expression of cyclin F/CCNF is an unfavorable prognostic factor that correlates with aggressive tumor phenotype and poor patient survival. Mechanistically, our bioinformatics analyses suggest that CCNF may act as an oncogene in ccRCC via promoting cancer cell proliferation and affecting the tumor immune microenvironment. Further investigation, including larger cohort and in vitro studies, is needed to describe molecular pathways altered along with CCNF overexpression.
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: https://www.cbioportal.org (accessed on 14 September 2022); https://xenabrowser.net/, (accessed on 14 September 2022); http://tnmplot.com (accessed on 28 November 2022); http://gdancik.github.io/shinyGEO/ (accessed on 30 November 2022); http://ualcan.path.uab.edu/ (accessed on 30 November 2022 and 23 March 2023); http://gent2.appex.kr/gent2/ (accessed on 29 November 2022); http://gepia2.cancer-pku.cn/ (accessed on 13 December 2022); http://kmplot.com/analysis (accessed on 13 December 2022); https://david.ncifcrf.gov (accessed on 23 March 2023). Own TMA_1 dataset used and analyzed during the current study are available from the corresponding author on reasonable request. A commercially purchased TMA_2 dataset is available online at https://www.tissuearray.com/ under the no. KD20810a.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Vasudev, N. S. et al. Challenges of early renal cancer detection: Symptom patterns and incidental diagnosis rate in a multicentre prospective UK cohort of patients presenting with suspected renal cancer. BMJ Open 10, e035938 (2020).
Moch, H., Cubilla, A. L., Humphrey, P. A., Reuter, V. E. & Ulbright, T. M. WHO Classification of Tumours of the Urinary System and Male Genital Organs (International Agency for Research on Cancer Press, 2016).
Xue, J. et al. Patterns of distant metastases in patients with clear cell renal cell carcinoma—A population-based analysis. Cancer Med. 10, 173–187 (2021).
Leibovich, B. C. et al. Histological subtype is an independent predictor of outcome for patients with renal cell carcinoma. J. Urol. 183, 1309–1315 (2010).
Noguchi, G. et al. Time-dependent change in relapse sites of renal cell carcinoma after curative surgery. Clin. Exp. Metastasis 35, 69–75 (2018).
Makhov, P. et al. Resistance to systemic therapies in clear cell renal cell carcinoma: Mechanisms and management strategies. Mol. Cancer Ther. 17, 1355–1364 (2018).
Kaldany, A. et al. Clinicopathological and survival analysis of clinically advanced papillary and chromophobe renal cell carcinoma. Urol. Oncol. 37, 727–734 (2019).
D’Angiolella, V., Esencay, M. & Pagano, M. A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis. Trends Cell Biol. 23, 135–140 (2013).
Klimaszewska-Wiśniewska, A. et al. Expression of genomic instability-related molecules: Cyclin F, RRM2 and SPDL1 and their prognostic significance in pancreatic adenocarcinoma. Cancers (Basel) 13, (2021).
Fu, J. et al. Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 104, 508–515 (2013).
Gagat, M., Krajewski, A., Grzanka, D. & Grzanka, A. Potential role of cyclin F mRNA expression in the survival of skin melanoma patients: Comprehensive analysis of the pathways altered due to cyclin F upregulation. Oncol. Rep. 40, 123–144 (2018).
Wang, X., Zhang, T., Zhang, S. & Shan, J. Prognostic values of F-box members in breast cancer: an online database analysis and literature review. Biosci. Rep. 39, (2019).
Zelong, Y. et al. Increased expression of Cyclin F in liver cancer predicts poor prognosis: A study based on TCGA database. Medicine (Baltimore) 100, e26623 (2021).
Ogłuszka, M., Orzechowska, M., Jędroszka, D., Witas, P. & Bednarek, A. K. Evaluate Cutpoints: Adaptable continuous data distribution system for determining survival in Kaplan–Meier estimator. Comput. Methods Progr. Biomed. 177, 133–139 (2019).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020).
Bartha, Á. & Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 22, 2622 (2021).
Dumas, J., Gargano, M. A. & Dancik, G. M. shinyGEO: A web-based application for analyzing gene expression omnibus datasets. Bioinformatics 32, 3679–3681 (2016).
Chandrashekar, D. S. et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 25, 18–27 (2022).
Park, S.-J., Yoon, B.-H., Kim, S.-K. & Kim, S.-Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genomics 12, 101 (2019).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560 (2019).
Lánczky, A. & Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23, e27633 (2021).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020).
Szklarczyk, D. et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Chin, C.-H. et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8, S11 (2014).
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 4, 2 (2003).
Sherman, B. T. et al. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Bindea, G. et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Combescure, C., Perneger, T. V., Weber, D. C., Daurès, J.-P. & Foucher, Y. Prognostic ROC curves: A method for representing the overall discriminative capacity of binary markers with right-censored time-to-event endpoints. Epidemiology 25, 103–109 (2014).
Lim, S. & Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 140, 3079–3093 (2013).
Wei, L. et al. CCNF is a potential pancancer biomarker and immunotherapy target. Front. Oncol. 13, 1109378 (2023).
Gao, X. et al. A comprehensive analysis of the prognostic, immunological and diagnostic role of CCNF in pan-cancer. J. Cancer 14, 2431–2442 (2023).
Li, X., Li, Y.-J., Wang, M.-J., Ou, K.-P. & Chen, Y.-Q. Inhibition of cyclin F promotes cellular senescence through cyclin-dependent kinase 1-mediated cell cycle regulation. Curr. Med. Sci. 43, 246–254 (2023).
Hu, F., Zeng, W. & Liu, X. A gene signature of survival prediction for kidney renal cell carcinoma by multi-omic data analysis. Int. J. Mol. Sci. 20, 5720 (2019).
Aran, D. et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 8, 1077 (2017).
Schaeffer, D. F. et al. Insulin-like growth factor 2 mRNA binding protein 3 (IGF2BP3) overexpression in pancreatic ductal adenocarcinoma correlates with poor survival. BMC Cancer 10, 59 (2010).
Klimaszewska-Wiśniewska, A. et al. Expression of genomic instability-related molecules: Cyclin F, RRM2 and SPDL1 and their prognostic significance in pancreatic adenocarcinoma. Cancers 13, 859 (2021).
Liu, Y. et al. Systematic analysis of the expression and prognosis relevance of FBXO family reveals the significance of FBXO1 in human breast cancer. Cancer Cell Int. 21, 130 (2021).
Krajewski, A. et al. Cyclin F downregulation affects epithelial-mesenchymal transition increasing proliferation and migration of the A-375 melanoma cell line. CMAR 12, 13085–13097 (2020).
Deshmukh, R. S., Sharma, S. & Das, S. Cyclin F-dependent degradation of RBPJ inhibits IDH1R132H-mediated tumorigenesis. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-18-1772 (2018).
Barabási, A.-L., Gulbahce, N. & Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 (2011).
Liu, X. D., Yao, D. W. & Xin, F. TTK contributes to tumor growth and metastasis of clear cell renal cell carcinoma by inducing cell proliferation and invasion. Neoplasma 66, 946–953 (2019).
Chen, Z. & Wang, L. The clinical significance of UBE2C gene in progression of renal cell carcinoma. Eur. J. Histochem. 65, 3196 (2021).
Peng, R., Wang, Y., Mao, L., Fang, F. & Guan, H. Identification of core genes involved in the metastasis of clear cell renal cell carcinoma. Cancer Manag. Res. 12, 13437–13449 (2020).
Deng, H., Gong, X., Ji, G., Li, C. & Cheng, S. KIF2C promotes clear cell renal cell carcinoma progression via activating JAK2/STAT3 signaling pathway. Mol. Cell Probes. 72, 101938 (2023).
Zhang, Z., Zhang, G. & Kong, C. FOXM1 participates in PLK1-regulated cell cycle progression in renal cell cancer cells. Oncol. Lett. 11, 2685–2691 (2016).
Wan, B., Huang, Y., Liu, B., Lu, L. & Lv, C. AURKB: A promising biomarker in clear cell renal cell carcinoma. PeerJ 7, e7718 (2019).
Li, H. et al. NCAPG promotes the proliferation of renal clear cell carcinoma via mediating with CDK1. Dis. Mark. 2022, 6758595 (2022).
Ikuerowo, S. O. et al. Alteration of subcellular and cellular expression patterns of cyclin B1 in renal cell carcinoma is significantly related to clinical progression and survival of patients. Int. J. Cancer 119, 867–874 (2006).
Hu, C. et al. NDC80 status pinpoints mitotic kinase inhibitors as emerging therapeutic options in clear cell renal cell carcinoma. iScience 26, 106531 (2023).
Luo, Y., Shi, Q., Wang, L., Li, S. & Xu, W. Transcription factor 19-mediated epigenetic regulation of FOXM1/AURKB axis contributes to proliferation in clear cell renal carcinoma cells. MedComm (2020) 4, e442 (2023).
Fang, K. et al. FOXM1/KIF20A axis promotes clear cell renal cell carcinoma progression via regulating EMT signaling and affects immunotherapy response. Heliyon 9, e22734 (2023).
Tang, A. et al. Aurora kinases: Novel therapy targets in cancers. Oncotarget 8, 23937–23954 (2017).
Xue, Y.-J. et al. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J. Transl. Med. 10, 200 (2012).
Acknowledgements
We would like to greatly thank to Dr. Christophe Combescure, from the University of Geneva, for his generous help with the analysis of the prognostic ROC curve. The study was supported by the research task within the framework of Basic Research Activity Program, Faculty of Medicine, Ludwik Rydygier Collegium Medicum in Bydgoszcz, Nicolaus Copernicus University in Toruń.
Author information
Authors and Affiliations
Contributions
A.K.W., A.K., and D.G. conceptualized and designed the study. J.D., prepared tissue macroarrays and performed the immunohistochemical staining. D.G., A.K.W., and M.K. evaluated the immunohistochemical staining. D.G. and M.K. acquired the study data. A.K.W. performed the statistical analysis, A.K.W. and M.K. interpreted and visualized the data. M.K., A.K.W., A.K., and K.B. wrote the manuscript text. J.D., M.G., and J.Z. revised the manuscript. A.K.W., D.G., and M.G. supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kwiatkowski, M., Krajewski, A., Durślewicz, J. et al. Overexpression of cyclin F/CCNF as an independent prognostic factor for poor survival in clear cell renal cell carcinoma. Sci Rep 14, 9280 (2024). https://doi.org/10.1038/s41598-024-59437-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-59437-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.