Abstract
Alternaria, a cosmopolitan fungal genus is a dominant member of the grapevine (Vitis vinifera) microbiome. Several Alternaria species are known to produce a variety of secondary metabolites, which are particularly relevant to plant protection and food safety in field crops. According to previous findings, the majority of Alternaria species inhabiting grapevine belong to Alternaria sect. Alternaria. However, the phylogenetic diversity and secondary metabolite production of the distinct Alternaria species has remained unclear. In this study, our aim was to examine the genetic and metabolic diversity of endophytic Alternaria isolates associated with the above-ground tissues of the grapevine. Altogether, 270 Alternaria isolates were collected from asymptomatic leaves and grape clusters of different grapevine varieties in the Eger wine region of Hungary. After analyses of the nuclear ribosomal DNA internal transcribed spacer (ITS) and RNA polymerase second largest subunit (rpb2) sequences, 170 isolates were chosen for further analyses. Sequences of the Alternaria major allergen gene (Alt a 1), endopolygalacturonase (endoPG), OPA10-2, and KOG1058 were also included in the phylogenetic analyses. Identification of secondary metabolites and metabolite profiling of the isolates were performed using high-performance liquid chromatography (HPLC)–high-resolution tandem mass spectrometry (HR-MS/MS). The multilocus phylogeny results revealed two distinct groups in grapevine, namely A. alternata and the A. arborescens species complex (AASC). Eight main metabolites were identified in all collected Alternaria isolates, regardless of their affiliation to the species and lineages. Multivariate analyses of untargeted metabolites found no clear separations; however, a partial least squares-discriminant analysis model was able to successfully discriminate between the metabolic datasets from isolates belonging to the AASC and A. alternata. By conducting univariate analysis based on the discriminant ability of the metabolites, we also identified several features exhibiting large and significant variation between A. alternata and the AASC. The separation of these groups may suggest functional differences, which may also play a role in the functioning of the plant microbiome.
Similar content being viewed by others
Introduction
Grapevine (V. vinifera) holds significant economic importance globally, and it is associated with a diverse microbiome. A fungal core microbiome of grapevine, independent of the season and region, has been reported from recent studies using both culture-dependent and culture-independent methods, confirming the predominance of the genera Alternaria, Aureobasidium, Botrytis, Cladosporium, Epicoccum, Fusarium, and Penicillium1,2,3,4,5,6,7,8,9,10,11. According to these studies, members of Alternaria sect. Alternaria are among the most common endophytes and constitute a dominant fungal consortium in different grapevine varieties. Alternaria is a biologically, morphologically, and ecologically diverse fungal genus, including cosmopolitan saprobes, endophytes, pathogens, and causal agents of post-harvest rots, producing substantial economic losses for different agronomic plants12. Alternaria spp. have been commonly identified based on conidial and colony characteristics, although morphological traits, especially of small-spored species, often overlap13. Due to this heterogeneity, the delineation of species is challenging. Recent advances in multilocus phylogeny have accelerated the identification of Alternaria species, and the genus has been reorganized into 27 sections based on sequences commonly used in the molecular phylogeny of other fungal genera, including nrDNA SSU, LSU, ITS, β-tubulin, tef1, calmodulin, actin, and expanded analyses of sequences of rpb2, endoPG, Alt a 1, gapdh, OPA1-3, OPA10-2, KOG1058, and KOG107714,15,16,17,18,19,20,21,22,23,24,25. Among the 27 phylogenetic sections, section Alternaria—closely related to sections Alternantherae and Porri—consists of the species complex A. arborescens (AASC) and 11 phylogenetic species (A. burnsii, A. tomato, A. jacinthicola, A. iridiaustralis, A. eichhorniae, A. betae-kenyensis, A. gaisen, A. alstroemeriae, A. longipes, A. gossypina, and A. alternata), from which A. alternata comprises 35 morphospecies23. This genus has gained increasing attention due to its ability to produce a broad spectrum of biologically active secondary metabolites with phytotoxic, cytotoxic, and antimicrobial properties26. Alternaria species produce more than 250 metabolites, mainly nitrogen-containing compounds, steroids, terpenoids, pyranones, quinones, and phenolics26. The major Alternaria toxins belong to the chemical groups of dibenzo-α-pyrones and cyclic tetrapeptides27. Based on chemical structure, Alternaria mycotoxins can be divided into three further classes: perylene (derivatives); tetramic acid derivatives; and the TA1, TA2, TB1, and TB2 toxins from A. alternata subspecies lycopersici28. Qualitative analysis of secondary metabolite production using high-resolution chromatographic techniques has been successfully used as a tool for the segregation and differentiation of Alternaria29,30,31,32,33,34,35,36. These studies have demonstrated that metabolite profiling can be informative in the grouping of large-spored Alternaria isolates but have revealed further ambiguity among small-spored Alternaria species.
Culture-independent methods provide a powerful tool for detailed grapevine microbiome studies; although, these methods tend to perform poorly in species-level identification in many fungal groups. The ITS region of the nuclear ribosomal DNA has been commonly used to characterize the diversity and composition of fungal communities in grapevine; however, the taxonomic resolution of this single locus is low in the case of Alternaria37. The ITS region alone is not adequate for species-level discrimination, and species belonging to Alternaria sect. Alternaria have been frequently identified in grapevine as A. alternata (e.g.10). Due to their potential taxonomic diversity with distinct functions and metabolite production, gaining information regarding the grapevine-colonizing species of Alternaria sect. Alternaria would provide important insights.
As our previous results have demonstrated the common presence of diverse Alternaria members in the wine grape microbiome10, we aimed in this study to reveal the diversity of Alternaria colonizing asymptomatic tissues of different cultivars of V. vinifera in a historical wine region of Hungary. Our main goal was to collect Alternaria isolates, identify the taxonomic identity of the isolates using multilocus molecular phylogenetic analyses, and determine whether there is a correspondence between these groups based on an untargeted chemical analysis of the secondary metabolites produced by those isolates.
Materials and methods
Isolation of fungal endophytes
Different parts (leaves, grape clusters, and berries) of several grapevine varieties (Vitis vinifera cv. Furmint, cv. Pinot Noir, cv. Merlot, cv. Leányka, cv. Chardonnay, and cv. Kadarka, as well as unidentified cultivars) were sampled between August and September of 2019 from seven vineyards (Nagy-Eged hegy, Hangács, Kőlyuk-tető, Rác-hegy, Cinege, Déllés, and Hajdú-hegy) in different localities of the Eger wine region of Hungary (Supplementary Table 2). The St. Andrea Winery and the Centre for Research and Development at Eszterházy Károly Catholic University granted authorization to collect plant material for our research on V. vinifera cv. Furmint, cv. Pinot Noir, cv. Merlot, cv. Leányka, cv. Chardonnay, cv. Kadarka and unidentified cultivars. Sample collection has complied with relevant institutional, national, and international guidelines and legislation. The region is a cool-climate district located in northeastern Hungary, in the southern foreland of the Bükk Mountains38. The 22,160-ha area has high oenological value, containing 6000 ha of vineyards. Rhyolite tuff and volcanic rocks are the characteristic bedrock. Variations of brown forest soils containing clay minerals in high quantity are dominant in the area39.
In each location, young leaves (fourth from the shoot tip), mature leaves, and grape clusters (separated into berries and rachis/pedicels) were collected and then stored in plastic bags at 4 °C for further processing within 24 h. Endophyte isolation was conducted as described by Knapp et al.10, with modifications. Briefly, samples were sliced into 0.5–1-cm segments, soaked in 30% H2O2 for 1 min, then soaked in 70% ethanol for 1 min, and washed in sterile tap water twice for 2–3 min. Surface-sterilized samples were transferred onto potato dextrose agar (PDA) media (VWR, Germany) and incubated at 25 °C. Within the following 1–7 days, mycelia growing from the plant tissues were transferred to new PDA plates.
Genomic DNA extraction and molecular characterization
Mycelia were harvested from pure cultures and disrupted and homogenized in a TissueLyser LT (QIAGEN, Germany). Genomic DNA extraction was performed using the NucleoSpin Plant II DNA Isolation Kit (MACHEREY-NAGEL, Germany), following the manufacturer’s instructions. The internal transcribed spacer region (ITS) of the nuclear ribosomal DNA and partial region of the RNA polymerase II second largest subunit (rpb2) were amplified with the primer pairs ITS1F, ITS440,41, and RPB2-6F and fRPB2-7cR42, respectively. Representative isolates identified by ITS and rpb2 as members of Alternaria sect. Alternaria were chosen for further, multilocus sequence analysis. The loci of the Alternaria major allergen gene (Alt a 1), endopolygalacturonase (endoPG), an approximately 800-bp partial sequence of an anonymous noncoding region (OPA10-2), and the eukaryotic orthologous group (KOG) protein locus (KOG1058) were amplified by polymerase chain reaction with the primer pairs Alt-for and Alt-rev16, PG3 and PG2b17, OPA10-2R and OPA10-2L17, and KOG1058F2 and KOG1058R223, respectively. Because amplification of Alt a 1 failed using the primer pairs Alt-for and Alt-rev, PCR was carried out with the modified primers Alt4-for and Alt4-rev43. Reactions were performed in a T100 Thermal Cycler (Bio-Rad, California, USA) in a 50-µL reaction mixture. Phire Hot Start II PCR Master Mix (Thermo Fisher Scientific, Massachusetts, USA) was used for each reaction. PCR conditions consisted of an initial denaturation step at 98 °C for 30 s, followed by 32 cycles of 98 °C for 5 s denaturation at 98 °C for 5 s; the optimal annealing temperature for each primer pair (see Supplementary Table 3) for 5 s; and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 3 min. PCR products were submitted to Eurofins Genomics GmbH for amplicon sequencing (Ebersberg, Germany). Sequence analysis and editing were performed with the PREGAP4 and GAP4 tools of the Staden software package44 and deposited in GenBank (OQ931049–OQ931220, OQ973480–OQ974171; Supplementary Table 1). To obtain preliminary identification of the isolates, ITS sequences were blasted against available sequences from public databases using BLASTN searches45.
Phylogenetic analyses
We combined and aligned the sequences of the different loci with those from representative taxa in GenBank using the online version of MAFFT 746 and following the E-INS-i method. The alignments were examined and edited in MEGA 747. Three multilocus datasets were used for molecular phylogenetic analyses of the isolates and reference strains of Alternaria sect. Alternaria 23 (Supplementary Table 1). For the first dataset, we used sequences of six sequenced loci (ITS, rpb2, Alt a 1, endoPG, OPA10-2, and KOG1058) from our isolates (Supplementary Fig. 1). In the second dataset, five loci (ITS, rpb2, Alt a 1, endoPG, and OPA10-2) from our strains and reference strains of Alternaria sect. Alternaria sequenced in this study and by Woudenberg et al.23 were analyzed (Fig. 1). The third analysis used all seven loci (ITS, rpb2, Alt a 1, endoPG, OPA10-2, tef1, and gapdh) from the taxa analyzed by Woudenberg et al.23, and our isolates were represented by the shared five loci (Supplementary Fig. 2). In the two later phylogenies, A. alternantherae, CBS 124,392, and A. perpunctulata CBS 115,267 served as an outgroup. In the case of the ITS, rpb2, Alt a 1, endoPG, OPA10-2, tef1, and gapdh datasets, the partitions were analyzed separately to examine differences in single-locus phylogenies (Supplementary Fig. 3). Bayesian inference analyses were performed with MRBAYES 3.1.248 using a GTR + G substitution model for the nucleotide partitions. Four Markov chains were run for 10,000,000 generations, sampling every 1,000 generations with a burn-in value set at 4,000 sampled trees. Maximum likelihood (ML) phylogenetic analysis was performed with the RAXMLGUI 1.3 implementation49,50. A GTR + G nucleotide substitution model was used for nucleotide partitions with ML estimation of base frequencies. ML bootstrapping (BS) analysis with 1,000 replicates was used to test the support of the branches. Phylogenetic trees were visualized and edited in MEGA 747.
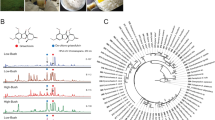
Phylogenetic tree of all Alternaria isolates collected from grapevine leaves and clusters and reference Alternaria strains from Woudenberg et al.23. The 50% majority rule consensus phylogram inferred from Bayesian analysis of the combined dataset of five loci (rpb2, ITS, Alt a 1, endoPG, OPA10-2). Bayesian posterior probabilities (≥ 0.90) are shown before slashes, ML bootstrap support (≥ 70) is shown after slashes. Isolates representing the lineages of A. alternata (violet) and A. arborescens species complex (AASC, dark green) in this study are shown in bold. Sequences were rooted to A. alternantherae, A. perpunctulata, A. solani, A. porri, A. tagetica, A. macrospora, A. pseudorostrata and A. dauci. Scale bar indicates 1 expected change per branch.
Metabolite extraction and identification
For metabolite profiling, 170 out of the 173 isolates (three A. alternata isolates were lost before these analyses) were subcultured simultaneously and grown in three replicates in Petri dishes (60 mm × 15 mm) on PDA medium (VWR, Hungary) at room temperature in the dark for 21 days. The entire culture-containing PDA medium and the fungal mycelium grown on it were lyophilized and pulverized. Solvents applied in the extraction analysis of metabolites, such as acetonitrile, distilled water, formic acid, and methanol (Reanal, Hungary), were all of the analytical reagent grade of the highest purity available. Aliquots of the powdered cultures (20.0 mg) were extracted with 5.0 mL of methanol in 25 mL screw-capped vials at 60 °C for 30 min. The insoluble, centrifuged material was subsequently re-extracted in the same way. The supernatants were combined to prepare 10.0 mL extracts.
To identify compounds present in extracts of Alternaria isolates, a Dionex Ultimate 3000 UHPLC system (3000RS diode array detector [DAD], TCC-3000RS column thermostat, HPG-3400RS pump, SRD-3400 solvent rack degasser, WPS-3000TRS autosampler), connected to an Orbitrap Q Exactive Focus Mass Spectrometer equipped with electrospray ionization (ESI) (Thermo Fischer Scientific, Waltham, MA, USA) was used. High-performance liquid chromatography (HPLC) separations were performed on a Kinetex C18 column (75 × 3 mm; 2.6 μm) (Phenomenex, USA). The eluent A consisted of 0.1% v/v formic acid in water, and eluent B, a mixture of acetonitrile and water in an 80:20, v/v ratio, containing 0.1% v/v formic acid. A linear gradient was applied with initial conditions of 20% B at 0.0 min, reaching 90% B at 12.0 min. The flow rate was maintained at 0.3 mL/min, the temperature was set to 25 °C, and a 5.0 μL volume was injected. The ESI source was operated in positive and negative ionization modes (switching mode). Fragmentations were performed by the data-independent acquisition method using isolation widths of 100–300 m/z, 295–500 m/z, 495–700 m/z, and 695–800 m/z and collision energies of 15, 30, and 45 eV. Operation parameters were optimized automatically by the built-in software as follows: spray voltage, 3500 V ( +); capillary temperature, 256 °C; sheath-, auxiliary-, and spare-gases (N2): 47.50, 11.25, and 2.25 arbitrary units, respectively. The resolutions of the full scans and MS/MS scans were 70,000 and 35,000, respectively. The full MS scanning range was 100–1000 m/z units. UV spectra were recorded between 250 and 600 nm, and UV chromatograms were plotted as summed signal intensities measured in this wavelength range.
Untargeted mass spectrometry (MS) data processing
Untargeted MS data processing was performed using MZmine 3 metabolomics software51. From each raw data file (.RAW) acquired in LC–MS analysis, positive and negative MS1 scan data were extracted into separate NetCDF files. As the MS data were recorded as centroid, a noise-filtering and mass-picking algorithm was used with an intensity threshold of 100,000, followed by a chromatogram building and deconvoluting step performed by the ADAP module52. The resulting feature lists (distinct RT-m/z pairs) were deisotoped and aligned between samples using the RANSAC algorithm53. Features not detected in all of the replicates of a sample were eliminated from the aligned feature list, which was then exported in .CSV format for further statistical analysis.
Statistical analyses of untargeted MS data
The positive and negative MS data were separately subjected to statistical analysis. In the metabolomic dataset, each feature’s abundance was expressed as peak area, and the data were centered and scaled to achieve zero means and unit variances. Principal component analysis (PCA) was performed as a preliminary analysis using the R package mixOmics54, followed by a partial least squares discriminant analysis (PLS-DA) using the same package. To evaluate the model’s performance, a three-fold cross-validation was performed, repeated ten times. Welch’s t-tests, Mann–Whitney–Wilcoxon tests, and receiver operating characteristic (ROC) analyses were conducted to assess the discriminant ability of each feature. Additionally, fold change and Z-factor were calculated using the R package imageHTS. Area under the ROC curve versus Welch’s p-value combined with a Z-factor plot was constructed based on the technique of Broadhurst et al.55. Heatmaps were generated using the ComplexHeatmap R package.
Results
Endophyte isolation
In this study, more than 450 plant samples were collected from different grapevine cultivars characteristic of the area of the Eger wine region of Hungary. We collected a total of 570 fungal endophytes after isolation from surface-sterilized young and mature healthy leaves and grape clusters of V. vinifera (Supplementary Table 2). Based on the analysis of the ITS sequences, the isolates belonged to several taxa, from which we identified 270 isolates representing species within Alternaria sect. Alternaria. Common members of the grapevine mycobiome were also detected, including Aureobasidium pullulans (28 isolates), Stemphylium vesicarium (12) and Botrytis cinerea (12), which were present in the highest number among the grapevine-associated isolates (data not shown). According to the groups resulting from phylogenetic analysis of the rpb2 sequences of the 270 Alternaria isolates, 173 representatives were chosen for further multilocus molecular phylogenetic analyses. From these isolates, four genomic loci (Alt a 1, endoPG, OPA10-2, and KOG1058) in addition to the ITS and rpb2 sequences were amplified and sequenced. Amplification of Alt a 1 and OPA10-2 was successful for all 173 isolates; 25 isolates failed for endoPG, and 52 failed for KOG1058 (Supplementary Table 1).
Molecular phylogeny
In the six-locus phylogeny of the 173 isolates, nearly half (74) of the isolates exhibited almost identical sequences and represented one major clade. One distinct, well-supported (B-PP = 1, ML-BS = 96) clade comprised 26 isolates (Supplementary Fig. S1). In the five-locus phylogeny, our isolates from grapevine were analyzed together with reference sequences23, which served as the basis for species delimitation and the concept of phylospecies in Alternaria sect. Alternaria (Fig. 1). All of our isolates grouped together with representative taxa of A. alternata and the AASC, and no other species of Alternaria sect. Alternaria, such as A. gossypina and A. iridiaustralis, were represented. Most of our isolates grouped with A. alternata lineages. The strongly supported (B-PP = 1, ML-BS = 65) AASC clade comprised the 26 isolates (representing the second group of grapevine-derived isolates) together with well-characterized AASC strains. In the seven-locus phylogeny of our isolates from grapevine and reference sequences23, we observed a similar arrangement of our isolates (Supplementary Fig. S2). Most of these grouped together with A. alternata strains, and 26 showed strong affiliation with the well-supported (B-PP = 0.99, ML-BS = 99) clade of AASC strains. The phylogenetic reconstructions from both the five-locus and seven-locus analyses produced better resolution among the isolates but resulted in similar topologies to the single-locus analysis, with separation of the studied isolates from the other nine phylogenetic species of Alternaria sect. Alternaria.
Single-locus trees
Although the single-locus phylogenies produced slight differences in species resolution in the case of the 11 species or species complexes in Alternaria sect. Alternaria23, most species could be distinguished consistently within these phylogenies (Supplementary Fig. S3). Based on the single-locus and multilocus trees, we could also assume that none of our isolates belonged to any of the nine clades representing the species A. alstroemeriae, A. betae-kenyensis, A. eichhorniae, A. gaisen, A. gossypina, A. iridiaustralis, A. jacinthicola, A. longipes, and A. tomato (Supplementary Fig. S3). Phylogeny based on rpb2 revealed higher variability among the isolates than analyses using ITS and revealed two distinct groups of the isolates, similarly to the multilocus tree. Analysis of 157 sequences of Alt a 1, 144 of endoPG, and 170 of OPA 10–2 resulted in assignment of the same two groups. The most appropriate locus for distinguishing AASC isolates from A. alternata isolates was OPA 10–2, as AASC sequences were separated (BS = 84) in the single-locus phylogeny of this region. Using rpb2 also resulted in a distinct clade with (BS = 73). In our analyses, none of the other loci revealed clearly separated clades for all reference sequences of the AASC (Supplementary Fig. S3).
Untargeted chemical profiling and metabolite identification
All 170 isolates of A. alternata and the AASC from grapevine were analyzed by HPLC coupled to a DAD and high-resolution tandem mass spectrometry (HR-MS/MS) detections for metabolic profiling. Untargeted MS data processing revealed the presence of 647 and 453 distinct molecular features using positive and negative ionization, respectively. PCA of these datasets showed no significant separation of A. alternata and the AASC, indicating that the major source of variation is not attributable to the grouping of A. alternata species and the AASC, as shown in the heatmaps (Fig. 2a,c). However, a PLS-DA model built with two components successfully discriminated between the two groups by chemical profile, suggesting a difference in the chemical composition of these groups (Fig. 2b,d). Univariate statistical analysis of each feature as a binary classifier (Supplementary Fig. 9) revealed that 75 and 54 metabolites in positive and negative mode, respectively, could discriminate between A. alternata and the AASC. Most of the discriminant metabolites appear to be more abundant in A. alternata, with the exception of three features with higher concentrations in the AASC.
The culture extracts contained eight Alternaria-specific compounds (Fig. 3), which could be identified using molecular formulas (Table 1) and UV spectra identical to those of known metabolites of Alternaria species. We detected alternarienonic acid, alternarian acid, altenuene, L-tenuazonic acid, altenusin, alternariol, 4-hydroxyalternariol methyl ether, and alternariol monomethyl ether (Supplementary Figs. S4–S7). All 170 isolates produced these main compounds, although at different concentrations. Although the average concentrations and variances of these metabolites were similar in A. alternata and the AASC (negative Z-prime and AUROC < 0.8), two of these, namely altenuene and L-tenuazonic acid, differed significantly (Bonferroni-corrected p < 0.007) between the two groups, and differences between certain clades were also present. Among these molecules, we observed a distinct pattern for L-tenuazonic acid (Supplementary Fig. 8).
High-performance liquid chromatography ultraviolet spectrophotometry (HPLC–UV) separations (a, b) of the extracts prepared from Alternaria alternata isolate vvmerl3ml5 (a) and A. arborescens species complex (AASC) isolate vvunid6yl6 (b), and the chemical structures of their main compounds 1 alternarienonic acid, 2 alternerian acid, 3 altenuene, 4 L-tenuazonic acid, 5 altenusin, 6 alternariol, 7 4-hydroxyalternariol methyl ether and 8 alternariol monomethyl ether (λ = 250–600 nm).
Discussion
Based on economic value and hectares cultivated, grapevine is one of the major crops worldwide56 and has been described as a host plant of highly diversified microbial communities by many authors in the recent decade. Using both culture-dependent and culture-independent methods, it has been shown that species of Alternaria sect. Alternaria and related Alternaria species are represented among the endophytic fungal communities inhabiting the healthy tissues of grapevine1,2,3,4,5,6,7,8,9,10,11. Through the combination of morphological characterization, multigene sequence analysis, and metabolite profiling, this taxonomically challenging genus has undergone several revisions15,16,17,18,19,20,21,22,23,24,25,29,30,31,32,33,34,35,36,57 in the decades since its first description58. A comprehensive morphological approach by Simmons57 resulted in the description of more than 270 Alternaria species and the division of the genus into subgeneric sections. Alternaria is currently divided into 27 sections, from which members of Alternaria sect. Alternaria have been identified in significant numbers in our previous work10 based on sequences of ITS and rpb2. In this study, we subjected endophytic Alternaria strains isolated from asymptomatic grapevine leaves and clusters to a combined analysis of five additional genomic loci and metabolite profiling for improved species resolution within the Alternaria section.
For multilocus analysis of Alternaria isolates, ITS, rpb2, Alt a 1, endoPG, OPA10-2, and KOG1058 were sequenced, the genomic sequences of these loci, along with gapdh and tef1, have been used previously to evaluate phylogenetic relationships among species in Alternaria sect. Alternaria23. Based on both single-locus and multilocus phylogeny, the Alternaria strains obtained from grapevine represented two distinct lineages within Alternaria sect. Alternaria, considered as A. alternata and the AASC. Despite taxonomic differences between A. alternata and the AASC, their known host range, biology, and growing characteristics are similar. This condition has been confirmed by the fact that many of our isolates belonging to A. alternata and the AASC originated from the same plant sample (e.g., vvchar5yl1 and vvchar5yl3, Supplementary Table 2). Our results are consistent with those reported by Lorenzini and Zapparoli59 and Tao et al.60, who found that grapevine-derived Alternaria isolates clearly divided into two major clusters: A. alternata and the AASC.
By combining RAPD-PCR, morphological characterization, and metabolite profiling, Polizzotto12 identified endophytic Alternaria strains originating from grapevine shoots as members of the AASC and the A. tenuissima species group; they were distinct from strains belonging to A. alternata. This outcome suggests that small-spored Alternaria species other than A. alternata and the AASC may also be present in grapevine; however, we only detected these two groups in our study. Tao et al.60 have described a new species (A. viniferae) isolated from pedicels and rachis and closely related to A. longipes, which is the other grapevine-associated Alternaria strain clustering with A. alternata and the AASC based on sequence data from the gpd and Alt a 1 genes. Phylogenetic analysis of ITS sequences clustered all Alternaria isolates obtained from withered grapes within a monophyletic clade, while intergenic spacer region (IGS)-RFLP profiles were congruent with those of A. alternata and the AASC, as reported by Lorenzini and Zapparoli59.
Based on the analysis of ITS, LSU, gapdh, TEF, and rpb2 sequences, Dissanayake et al.6 identified all Alternaria isolates from grapevine stems as A. alternata sensu stricto. These results support our findings that not all genomic loci that have been successfully applied in studies of other fungal genera provide sufficient resolution among species in Alternaria sect. Alternaria but that the combination of informative loci in multigene phylogeny could assist with the more precise delimitation of species.
Alternaria isolates from symptomatic pomegranate fruits have also been identified as A. alternata and the AASC based on ITS, tef1, gapdh, and OPA10-2 sequences. Despite the morphological and genetic differences within these fungi, all tested isolates of both A. alternata and the AASC had a similar effect and induced similar symptoms of heart rot in pomegranate fruit61. By combining Alt a 1, gapdh, tef1, and tub gene sequences, Somma et al.62 also successfully distinguished A. alternata from AASC strains isolated from wheat kernels and found Alt a 1 to be the most informative locus according to the percentage of polymorphic sites. The two species were also found together by Fontaine63, who performed multilocus sequence analysis targeting the endoPG, Alt a 1, and OPA10-2 regions among Alternaria isolates from leaves and fruits of apple samples affected by Alternaria leaf blotch (ALB) and Alternaria fruit spot (AFS). These two Alternaria taxa are the major cause of ALB and AFS64 but colonize grapevine tissues without any visible symptoms.
The presence of Alternaria species in agronomic plants is frequently connected with the accumulation of secondary metabolites representing a wide variety of biological activities. The prevalence of these species is of high importance in food safety and plant pathology, as many of the produced metabolites have been classified as mycotoxins or phytotoxins34. In recent years, several substrates, such as cereals, fruits, and derived products intended for human or animal consumption, have been analyzed for the presence of Alternaria metabolites, focusing mainly on mycotoxins27. However, toxicity and modes of action have not been elucidated in detail for all substances. Eight known fungal secondary metabolites were identified in the grapevine-associated Alternaria isolates that we investigated, namely alternarienonic acid, alternarian acid, altenuene, L-tenuazonic acid, altenusin, alternariol, 4-hydroxyalternariol methyl ether, and alternariol methyl ether (Fig. 3), of which altenuene, L-tenuazonic acid, altenusin, alternariol, and alternariol methyl ether have previously been reported in Alternaria isolated from different grapevine organs65,66,67,68. Alternarienonic acid was identified first in cultures of endophytic Alternaria spp. isolated from Polygonum senegalense69 and later from the mangrove plant Sonneratia alba70 as well as blueberries, walnuts, tomatoes, and wheat34. Alternarian acid was identified in cultured A. mali isolated from naturally infected tobacco71. Toxicity of the dibenzopyrone derivative altenuene was first reported against different bacteria72. Several Alternaria species also produce various phytotoxins that are both host-specific and non-host-specific. Tenuazonic acid belongs to the tetramic acid derivative group of Alternaria metabolites and has been widely detected in several agronomic plants, such as cottonseed and bolls73. It has been described as a non-host-specific, nitrogen-containing phytotoxin inhibiting photophosphorylation74 and was found to have antimicrobial properties against Mycobacterium tuberculosis75. Tenuazonic acid has also been linked to several adverse effects on animal species such as mice, chickens, and dogs76,77 and has been found to inhibit protein biosynthesis in rat liver cells and Ehrlich ascites by suppressing the release of nascent proteins from the ribosome78. It has been detected along with alternariol and alternariol monomethyl ether in food products such as ice wines79, cornflakes80, sunflower flour81, and tomato puree82. Tenuazonic acid is also produced by the fungi Pyricularia oryzae and Phoma sorghina83,84. Altenusin is a biphenyl derivative with antioxidant activity and the capability to inhibit various enzymes of Paracoccidioides brasiliensis and Schizosaccharomyces pombe85. Altenusin isolated from the plant Trixis vauthieri has been found to act as a potential chemotherapeutic agent to treat leishmaniasis and trypanosomiasis by inhibiting the drug target trypanothione reductase86. Altenuene, alternariol, alternariol methyl ether, and 4-hydroxyalternariol methyl ether are dibenzo-α-pyrones possessing antibacterial and antioxidant effects. Like tenuazonic acid, alternariol is a non-host-specific phytotoxin28. Furthermore, alternariol induces lipid peroxidation in the epithelium of the human fetal esophagus in vitro87 and has been reported to exhibit genotoxic activity by inducing DNA strand breaks in cultured mammalian cells88,89,90. Alternariol methyl ether can also inhibit type II DNA topoisomerases in human colon adenocarcinoma cells91. Alternariol is secreted by other fungi in addition to Alternaria species, including Stagonospora nodorum92 and Phomopsis sp.93. The compound 4-hydroxyalternariol methyl ether and alternariol from endophytic Alternaria sp. obtained from the medicinal plant Salvia miltiorrhiza have shown antibacterial activities against six pathogenic bacteria94. Of note, 4-hydroxyalternariol methyl ether is produced not only by Alternaria species but also by endolichenic fungal strains of Nigrospora sp.95.
The multivariate analyses of the untargeted metabolite characterization we performed found no clear separation among Alternaria species in the metabolomic space. However, a PLS-DA model was able to successfully discriminate between the metabolic data from isolates belonging to the AASC and the remaining A. alternata isolates. By conducting univariate analysis based on the discriminant ability of the metabolites, we also identified several features with large and significant variation between A. alternata and the AASC. Although well-known Alternaria metabolites were detected in all isolates at similar concentrations, the amounts of two chemicals, altenuene and L-tenuazonic acid, differed significantly between A. alternata and the AASC.
The distinct grouping of the AASC among A. alternata, revealed previously and supported by our present molecular phylogenetic analyses, is supported by the data obtained from the untargeted metabolic study. This separation of groups may suggest functional differences, which could explain how isolates of the two groups could be found even in the same small tissue samples. Further studies are needed to understand the role of this metabolic and potentially functional diversity in the functioning of the plant microbiome.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. DNA sequences of A. alternata and A. arborescens species complex used in this study were deposited in GenBank (OQ931049–OQ931220, OQ973480–OQ974171).
References
González, V. & Tello, M. L. The endophytic mycota associated with Vitis vinifera in Central Spain. Fungal. Divers. 47, 29–42 (2011).
Pancher, M. et al. Fungal endophytic communities in grapevines (Vitis vinifera L.) respond to crop management. Appl. Environ. Microbiol. 78, 4308–4317 (2012).
Pinto, C. et al. Unravelling the diversity of grapevine microbiome. PLoS ONE 9, e85622. https://doi.org/10.1128/AEM.07655-11 (2014).
Setati, M. E., Jacobson, D. & Bauer, F. F. Sequence-based analysis of the Vitis vinifera L. cv cabernet sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front. Microbiol. 6, 1358. https://doi.org/10.3389/fmicb.2015.01358 (2015).
Kernaghan, G., Mayerhofer, M. & Griffin, A. Fungal endophytes of wild and hybrid Vitis leaves and their potential for vineyard biocontrol. Can. J. Microbiol. 63, 583–595 (2017).
Dissanayake, A. J. et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Divers. 90, 85–107 (2018).
Jayawardena, R. S. et al. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. 90, 1–84. https://doi.org/10.1007/s13225-018-0398-4 (2018).
Wei, Y. et al. High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS ONE 13, e0193097. https://doi.org/10.1371/journal.pone.0193097 (2018).
Liu, D. & Howell, K. Community succession of the grapevine fungal microbiome in the annual growth cycle. Environ. Microbiol. 23, 1842–1857 (2021).
Knapp, D. G. et al. Above-ground parts of white grapevine Vitis vinifera cv. Furmint share core members of the fungal microbiome. Environ. Microbiol. Rep. 13, 509–520 (2021).
Gramaje, D. et al. Exploring the temporal dynamics of the fungal microbiome in rootstocks, the lesser-known half of the grapevine crop. J. Fungi 8, 421. https://doi.org/10.3390/jof8050421 (2022).
Polizzotto, R. et al. A polyphasic approach for the characterization of endophytic Alternaria strains isolated from grapevines. J. Microbiol. Methods 88, 162–171 (2012).
Zhu, X. Q. & Xiao, C. L. Phylogenetic, morphological, and pathogenic characterization of Alternaria species associated with fruit rot of blueberry in California. Phytopathology 105, 1555–1567 (2015).
Pryor, B. M. & Gilbertson, R. L. Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycol. Res. 104, 1312–1321 (2000).
Peever, T. L., Su, G., Carpenter-Boggs, L. & Timmer, L. W. Molecular systematics of citrus associated Alternaria species. Mycologia 96, 119–134 (2004).
Hong, S. G., Cramer, R. A., Lawrence, C. B. & Pryor, B. M. Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 42, 119–129 (2005).
Andrew, M., Peever, T. L. & Pryor, B. M. An expanded multi-locus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101, 95–109 (2009).
Lawrence, D. P., Gannibal, P. B., Peever, T. L. & Pryor, B. M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 105, 530–546 (2013).
Lawrence, D. P., Gannibal, P. B., Dugan, F. M. & Pryor, B. M. Characterization of Alternaria isolates from the Infectoria species-group and a new taxon from Arrhenatherum, Pseudoalternaria arrhenatheria sp. nov. Mycol. Prog. 13, 257–276. https://doi.org/10.1007/s11557-013-0910-x (2014).
Lawrence, D. P., Rotondo, F. & Gannibal, P. B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 15, 3. https://doi.org/10.1007/s11557-015-1144-x (2016).
Woudenberg, J., Groenewald, J., Binder, M. & Crous, P. Alternaria redefined. Stud. Mycol. 75, 171–212 (2013).
Woudenberg, J., Truter, M., Groenewald, J. & Crous, P. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 79, 1–47 (2014).
Woudenberg, J. H. C., Seidl, M. F. & Groenewald, J. Z. Alternaria section Alternaria: Species, formae speciales or pathotypes?. Stud. Mycol. 82, 1–21 (2015).
Armitage, A. D. et al. Discrete lineages within Alternaria alternata species group: Identification using new highly variable loci and support from morphological characters. Fungal Biol. 119, 994–1006 (2015).
Armitage, A. D. et al. Genomics evolutionary history and diagnostics of the Alternaria alternata species group including apple and Asian pear pathotypes. Front Microbiol. 10, 3124. https://doi.org/10.3389/fmicb.2019.03124 (2020).
Lou, J., Fu, L., Peng, Y. & Zhou, L. Metabolites from Alternaria fungi and their bioactivities. Molecules 18, 5891–5935 (2013).
Escrivá, L., Oueslati, S., Font, G. & Manyes, L. Alternaria mycotoxins in food and feed: An overview. J. Food Qual. 5, 1–20. https://doi.org/10.1155/2017/1569748 (2017).
Meena, M. & Samal, S. Alternaria host-specific (HSTs) toxins: An overview of chemical characterization, target sites, regulation and their toxic effects. Toxicol. Rep. 6, 745–758. https://doi.org/10.1016/j.toxrep.2019.06.021 (2019).
Andersen, B., Krøger, E. & Roberts, R. G. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol. Res. 105, 291–299 (2001).
Andersen, B., Krøger, E. & Roberts, R. G. Chemical and morphological segregation of Alternaria arborescens, A. infectoria and A. tenuissima species groups. Mycol. Res. 106, 170–182 (2002).
Andersen, B., Hansen, M. E. & Smedsgaard, J. Automated and unbiased image analyses as tools in phenotypic classification of small-spored Alternaria spp. Phytopathology 95, 1021–1029 (2005).
Andersen, B., Dongo, A. & Pryor, B. M. Secondary metabolite profiling of Alternaria dauci, A. porri, A. solani and A. tomatophila. Mycol. Res. 112, 241–250 (2008).
Andersen, B., Sørensen, J. L., Nielsen, K. F., van den Ende, B. G. & de Hoog, S. A polyphasic approach to the taxonomy of the Alternaria infectoria species–group. Genet. Biol. 46, 642–656 (2009).
Andersen, B., Nielsen, K. F., Fernández Pinto, V. & Patriarca, A. Characterization of Alternaria strains from Argentinean blueberry, tomato, walnut and wheat. Int. J. Food Microbiol. 196, 1–10 (2015).
Andersen, B. & Tharne, U. Differentiation of Alternaria infectoria and Alternaria alternata based on morphology, metabolite profiles, and cultural characteristics. Can. J. Microbiol. 42, 685–689 (1996).
Patriarca, A., Cabral, L. C., Pavicich, M. A., Nielsen, K. F. & Andersen, B. Secondary metabolite profiles of small-spored Alternaria support the new phylogenetic organization of the genus. Int. J. Food Microbiol. 291, 135–143 (2019).
Lücking, R. et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 11, 14. https://doi.org/10.1186/s43008-020-00033-z (2020).
Nagy, R. et al. Evaluation of the relationship between soil erosion and the mineral composition of the soil: A case study from a cool climate wine region of Hungary. Carpathian J. Earth Environ. Sci. 7, 223–230 (2012).
Bálo, B. et al. Focus on terroir studies in the Eger wine region of Hungary. In IVES Conference Series, Terroir. https://ives-openscience.eu/5128/ (2014).
Gardes, M. & Bruns, T. D. ITS primers with enhanced specifity for Basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993).
White, T. J., Bruns, T., Lee, S. & Taylor, J. W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A. et al.) 315–322 (Academic Press, 1990).
Liu, Y. J., Whelen, S. & Hall, B. D. Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808 (1999).
Lawrence, D. P., Park, M. S. & Pryor, B. M. Nimbya and Embellisia revisited, with nov. comb for Alternaria celosiae and A. perpunctulata. Mycol. Prog. 11, 799–815 (2012).
Staden, R., Beal, K. F. & Bonfield, J. K. The Staden package, 1998. Methods Mol. Biol. 132, 115–130 (2000).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 572–1574 (2003).
Silvestro, D. & Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337 (2012).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Schmid, R. et al. Integrative analysis of multimodal mass spectrometry data in MZmine 3. Nat. Biotechnol. 41, 447–449. https://doi.org/10.1038/s41587-023-01690-2 (2023).
Myers, O. D., Sumner, S. J., Li, S., Barnes, S. & Du, X. One step forward for reducing false positive and false negative compound identifications from mass spectrometry metabolomics data: New algorithms for constructing extracted ion chromatograms and detecting chromatographic peaks. Anal. Chem. 89, 8696–8703 (2017).
Fischler, M. A. & Bolles, R. C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 24, 381–395 (1981).
Rohart, F., Gautier, B., Singh, A. & LêCao, K. A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752. https://doi.org/10.1371/journal.pcbi.1005752 (2017).
Broadhurst, D. I. & Kell, D. B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2, 171–196 (2006).
Torregrosa, L., Vialet, S., Adivèze, A., Iocco-Corena, P. & Thomas, M. R. Grapevine (Vitis vinifera L.). Methods Mol. Biol. 1224, 177–194 (2015).
Simmons, E. G. Alternaria—An Identification Manual (CBS Fungal Biodiversity Centre, 2007).
von Esenbeck, N. & Daniel, C. G. Das System der Pilze und Schwämme. (1816).
Lorenzini, M. & Zapparoli, G. Characterization and pathogenicity of Alternaria spp. strains associated with grape bunch rot during post-harvest withering. Int. J. Food Microbiol. 186, 1–5 (2014).
Tao, W. C. et al. A new Alternaria species from grapevine in China. Mycol. Prog. 13, 999. https://doi.org/10.1007/s11557-014-0999-6 (2014).
Aloi, F. et al. Characterization of Alternaria species associated with heart rot of pomegranate fruit. J. Fungi 7, 172. https://doi.org/10.3390/jof7030172 (2021).
Somma, S., Amatulli, M. T., Masiello, M., Moretti, A. & Logrieco, A. F. Alternaria species associated to wheat black point identified through a multi-locus sequence approach. Int. J. Food Microbiol. 293, 34–43 (2019).
Fontaine, K. et al. Identification and pathogenicity of Alternaria species associated with leaf blotch disease and premature defoliation in French apple orchards. PeerJ 9, e12496. https://doi.org/10.7717/peerj.12496 (2021).
Rotondo, F., Collina, M., Brunelli, A. & Pryor, B. M. Comparison of Alternaria spp. collected in Italy from apple with A. mali and other AM-toxin producing strains. Phytopathology 102, 1130–1142 (2012).
Prendes, L. P., Merín, M. G., Andreoni, M. A., Ramirez, M. L. & Morata de Ambrosini, V. I. Mycobiota and toxicogenic Alternaria spp. strains in Malbec wine grapes from DOC San Rafael, Mendoza, Argentina. Food Control 57, 122–128 (2015).
Prendes, L. P. et al. Water activity and temperature effects on growth and mycotoxin production by Alternaria alternata strains isolated from Malbec wine grapes. J. Appl. Microbiol. 122, 481–492 (2017).
Tančinová, D., Mašková, Z., Rybárik, Ľ, Felšöciová, S. & Císarová, M. Colonization of grapes berries by Alternaria sp. and their ability to produce mycotoxins. Potravinarstvo 10, 7–13 (2016).
Stranska, M. et al. Fungal endophytes of Vitis vinifera—Plant growth promoters or potentially toxinogenic agents? Toxins 14, 66. https://doi.org/10.3390/toxins14020066 (2022).
Aly, A. H. et al. Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detection in its host plant Polygonum senegalense. J. Nat. Prod. 71, 972–980 (2008).
Kjer, J. et al. Xanalteric acids I and II and related phenolic compounds from an endophytic Alternaria sp. isolated from the mangrove plant Sonneratia alba. J. Nat. Prod. 72, 2053–2057 (2009).
Chadwick, D. J., Easton, I. W. & Johnstone, R. A. W. Fungal metabolites part 9. Isolation and x ray structure determination of alternarian acid from Alternaria mali. Tetrahedron 40, 2451–2456 (1984).
Pero, R. W., Owens, R. G., Dale, S. W. & Harvan, D. Isolation and identification of a new toxin, altenuene, from the fungus Alternaria tenuis. Biochim. Biophys. Acta 230, 170–179 (1971).
Davis, N. D., Diener, U. L. & Morgan-Jones, G. Tenuazonic acid production by Alternaria alternata and Alternaria tenuissima isolated from cotton. Appl. Environ. Microbiol. 2, 155–157 (1977).
Liu, Y.-X., Xu, X.-M., Dai, X.-B. & Qiang, S. Alternaria alternata crofton-weed toxin: a natural inhibitor of photosystem II in Chlamydomonas reinhardtii thylakoids. J. Agric. Food Chem. 55, 5180–5185 (2007).
Sonaimuthu, V. et al. Tenuazonic acid: A promising antitubercular principle from Alternaria alternata. Microbiol. Res. 2, 63–65 (2010).
Griffin, G. F. & Chu, F. S. Toxicity of the Alternaria metabolites alternariol, alternariol monomethyl ether, altenuene and tenuazonic acid in the chicken embryo assay. Appl. Environ. Microbiol. 46, 1420–1422 (1983).
Ostry, V. Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 1, 175–188 (2008).
Shigeura, H. T. & Gordon, C. N. The biological activity of tenuazonic acid. Biochemistry 2, 1132–1137 (1963).
Abramson, D., Delaquis, P. & Smith, D. Assessment of ochratoxin A and tenuazonic acid in Canadian ice-wines. Mycotoxin Res. 23, 147–151 (2007).
Aresta, A., Cioffi, N., Palmisano, F. & Zambonin, C. G. Simultaneous determination of ochratoxin A and cyclopiazonic, mycophenolic, and tenuazonic acids in cornflakes by solid phase microextraction coupled to high-performance liquid chromatography. J. Agric. Food Chem. 51, 5232–5237 (2003).
Combina, M. et al. Effect of heat treatments on stability of alternariol, alternariol monomethyl ether and tenuazonic acid in sunflower flour. Mycotoxin Res. 15, 33–38 (1999).
Terminiello, L., Patriarca, A., Pose, G. & Fernandez Pinto, V. Occurrence of alternariol, alternariol monomethyl ether and tenuazonic acid in Argentinean tomato puree. Mycotoxin Res. 22, 236–240 (2006).
Iwasaki, S., Muro, H., Nozoe, S., Okuda, S. & Sato, Z. Isolation of 3,4–dihydro–3,4,8–trihydroxy–2(2H)–naphthalenone and tenuazonic acid from Pyricularia oryzae cavara. Tetrahedron Lett. 1, 13–16 (1972).
Steyn, P. S. & Rabie, C. J. Characterisation of magnesium and calcium tenuazonate from Phoma sorghina. Phytochemistry 15, 1977–1979 (1976).
Johann, S. Antifungal activity of altenusin isolated from the endophytic fungus Alternaria sp. against the pathogenic fungus Paracoccidioides brasiliensis. Rev. Iberoam Micol. 29, 205–209 (2012).
Cota, B. B. Altenusin, a biphenyl isolated from the endophytic fungus Alternaria sp., inhibits trypanothione reductase from Trypanosoma cruzi. FEMS Microbiol. Lett. 285, 177–182 (2008).
Liu, G. T. Etiological role of Alternaria alternata in human esophageal cancer. Chin. Med. J. (Engl) 105, 394–400 (1992).
Brugger, E.-M. et al. Mutagenicity of the mycotoxin alternariol in cultured mammalian cells. Toxicol. Lett. 164, 221–230 (2006).
Pfeiffer, E., Eschbach, S. & Metzler, M. Alternaria toxins: DNA strand-breaking activity in mammalian cells in vitro. Mycotoxin Res. 23, 152–157 (2007).
Fehr, M. Alternariol acts as a topoisomerase poison, preferentially affecting the II alpha isoform. Mol. Nutr. Food Res. 53, 441–451 (2009).
Jarolim, K. et al. Dual effectiveness of Alternaria but not Fusarium mycotoxins against human topoisomerase II and bacterial gyrase. Arch. Toxicol. 91, 2007–2016 (2016).
Tan, K. C., Trengove, R. D., Maker, G. L., Oliver, R. P. & Solomon, P. S. Metabolite profiling identifies the mycotoxin in alternariol in the pathogen Stagonospora nodorum. Metabolomics 5, 330–335 (2009).
Abreu, L. M. Chemical and molecular characterization of Phomopsis and Cytospora-like endophytes from different host plants in Brazil. Fungal Biol. 116, 249–260 (2012).
Tian, J. Dibenzo-α-pyrones from the endophytic fungus Alternaria sp. Samif01: Isolation, structure elucidation, and their antibacterial and antioxidant activities. Nat. Prod. Res. 31, 387–396 (2017).
He, J. W. et al. (2012) Heptaketides with antiviral activity from three endolichenic fungal strains Nigrospora sp., Alternaria sp. and Phialophora sp. Fitoterapia 83, 1087–109 (2012).
Acknowledgements
This work was supported by the National Research, Development and Innovation Office, Hungary (Grants: VEKOP-2.3.3-15-2017-00020, OTKA NKFIH K-135712, OTKA NKFIH K-143453, ELTE Institutional Excellence Program 2020 -TKP2020-IKA-05, Diagnostics and Therapy 2, TKP2021-NKTA-16), the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (G. Tóth). The financial support from Bolyai+ New National Excellence Program of the Ministry for Innovation and Technology is highly appreciated (G. Tóth).
Funding
Open access funding provided by Eötvös Loránd University.
Author information
Authors and Affiliations
Contributions
G.M.K., D.G.K. and A.M. conceptualized and designed the research. A.M. and D.G.K. conducted the samplings, isolations and phylogenetic identification; G.T., M.L., and I.B. carried out metabolic analyses and evaluation; G.M.K. contributed to those. A.M., D.G.K., G.M.K., M.L., T.G. and B.I. wrote the manuscript. G.M.K., K.Z.V. and D.G.K. supervised and evaluated all stages of the research. All authors edited and commented on the first version of the manuscript and read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molnár, A., Knapp, D.G., Lovas, M. et al. Untargeted metabolomic analyses support the main phylogenetic groups of the common plant-associated Alternaria fungi isolated from grapevine (Vitis vinifera). Sci Rep 13, 19298 (2023). https://doi.org/10.1038/s41598-023-46020-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-46020-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.