Abstract
Oral squamous cell carcinoma (OSCC) is amongst the most common malignancies, with an estimated incidence of 377,000 and 177,000 deaths worldwide. The interval between the onset of symptoms and the start of adequate treatment is directly related to tumor stage and 5-year-survival rates of patients. Early detection is therefore crucial for efficient cancer therapy. This study aims to detect OSCC on clinical photographs (CP) automatically. 1406 CP(s) were manually annotated and labeled as a reference. A deep-learning approach based on Swin-Transformer was trained and validated on 1265 CP(s). Subsequently, the trained algorithm was applied to a test set consisting of 141 CP(s). The classification accuracy and the area-under-the-curve (AUC) were calculated. The proposed method achieved a classification accuracy of 0.986 and an AUC of 0.99 for classifying OSCC on clinical photographs. Deep learning-based assistance of clinicians may raise the rate of early detection of oral cancer and hence the survival rate and quality of life of patients.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) is among the most common malignancies worldwide, with a reported incidence of 377,713 and 117,757 deaths in 20201. The five-year survival rate is over 80% in the early stages, decreasing to < 30% for advanced disease. More than 60% of the OSCCs are diagnosed at an advanced stage with high morbidity and mortality2,3,4. The incidence and mortality rates underline the importance of oral cancer screening programs to improve early detection and therapeutic success5,6,7.
Although the golden standard is pathologically proven, early detection can be achieved visually as OSCCs start superficially from squamous cell metaplasia. Nonetheless, the diagnostic accuracy of primary health care professionals is limited, with a sensitivity of 57.8% and a specificity of between 31 and 53%8,9. The lack of adequate training, substantial heterogeneity, and the lack of experience impede an effective diagnosis by primary health care professionals6,10. An automated assistance system may improve the diagnostic accuracy, allowing a more reliable and accurate assessment of the oral cavity, especially in the hands of less experienced professionals.
With advancements in artificial intelligence, deep learning algorithms have been adopted in computer-aided detection and diagnosis (CAD). Mainly convolutional neural networks (CNN) have emerged as the state-of-the-art approach to medical image analysis. CNN's utilize convolutional kernels with small perceptual fields to extract features via weight sharing and local connectivity11. Recently, transformers have been introduced as an alternative approach to CNNs. Transformers are based on an attention mechanism that efficiently estimates each pixel-pair interplay12.
In oral and maxillofacial surgery, few studies have explored the capability of CNNs to automatically classify OSCC on clinical photographs. These studies addressed the classification and detection of oral potentially malignant diseases13 and oral cancer lesions5,14,15 using YOLOv5, ResNet-152, DensNet-161, Inception-v4 and EfficientNet-b4, respectively.
However, none of the studies has explored the accuracy of vision transformers for classifying OSCC. This study aims to develop an automated oral cancer screening system using vision transformers as a fundamental basis for a timely and accurate referral system.
Material and methods
Data
In the present study, 1406 clinical photographs (CPs) were randomly collected from the Department of Oral and Maxillofacial Surgery, Charité - Universitätsmedizin Berlin, Germany (mean age of 60.8 years, age range of 15–90 years). The photographs were acquired with single lens reflex (SLR) cameras with varying light exposures. The image resolution was a minimum of 72 dpi. CPs with masked lesions or foreign bodies (e.g., mirrors or tongue depressors) were excluded from further analyses as described in a previous study16. All image data were anonymized and de-identified before analysis. Informed consent for the analysis of data was obtained from all patients or their guardians in case of age below 18 years. This study has been conducted in accordance with the code of ethics of the World Medical Association (Declaration of Helsinki). The approval of this study was granted by the Institutional Review Board, the Ethics Committee of Charité – Universitätsmedizin Berlin (EA2/089/22).
Data classification
Different clinicians verified all CPs based on electronic medical records (EMR). CPs with OSCCs needed to be biopsy-proven. All CPs were subsequently reviewed and revised by three clinicians (RG, DT, TF). The three reviewers have at least five years of clinical experience. Each clinician was instructed and calibrated in the verification task using a standardized protocol before the selection and reviewing process. The final dataset consisted of 703 CPs of OSCC and 703 CPs of normal oral mucosae (Table 1).
The normal tissue dataset comprised photographs of the oral cavity without premalignant oral mucosal lesions or oral cancer. A further selection of the dataset to exclude possible anatomical variations or inflammatory conditions of the gingiva or mucosa was not performed.
The OSCC training dataset contained images of various tumor stages, including Tis (1%), T1 (28.9%), T2 (27.2%), T3 (16.3%), T4 (21.6%) and unknown tumor stages (5%). The locations were tongue (36.9%), floor of mouth (29.6%), maxilla (3.2%), mandible (15.8%), buccal mucosa (13.6%), palate (0.7%) and oropharynx (0.3%). The test and validation data sets contained a comparable distribution of tumor stages and locations with a maximum deviation of 10% from the training dataset.
The model
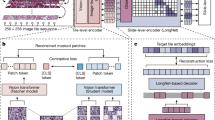
The Swin-Transformer was used in this study17. This transformer is characterized by its shift of the window partition between consecutive self-attention layers. The shifted windows connect with preceding layers’ window, increasing the modelling power efficiently. The employed model is shown in Fig. 1.
Model training
The annotated were randomly divided into 3 sets of CPs, 1124 for training, 141 for validation and 141 for testing. The validation set was used to evaluate the model performance during training, while the hold-out test set was used to evaluate the model performance after training.
The Swin-Transformer was pre-trained on the ImageNet dataset and optimized using a stochastic gradient descent with a learning rate of 5 × 10–3, a momentum of 0.9 and a weight decay of 1 × 10–4. No gradient clipping was applied. The model was implemented in PyTorch 1.11.0 and trained on a 12 GB NVIDIA TITAN V GPU. Model training was previously described in a study on caries detection radiographs16.
Statistical analysis
The transformer predictions on the test set were compared to the histopathological ground truth. Classification metrics are reported as follows for the test set: accuracy \(= \frac{TP+TN}{TP+TN+FP+FN}\), positive predictive value \(= \frac{TP}{TP+FP}\), F1-score \(= \frac{2TP}{2TP+FP+FN}\) , sensitivity \(= \frac{TP}{TP+FN}\) , specificity \(= \frac{TN}{TN+FP}\) , negative predictive value \(= \frac{TN}{TN+FN}\). TP, TN, FP, and FN denote true positives, true negatives, false positives, and false negatives, respectively. Furthermore, the area-under-the-curve-receiver-operating-characteristics-curve (AUC) and confusion matrix are presented. Gradient-weighted Class Activation Mapping (Grad-CAM), a class-discriminative localization technique was applied, to generate visual explanations highlighting the important regions on CPs for classifying OSCC. Statistical analysis was performed as in a previous study16.
Results
Table 2 summarizes the classification performance of the Swin-Transformer on the test set, including the accuracy, positive predictive value, sensitivity, specificity and negative predictive value. The classification accuracy was 98,6%. The model achieved an AUC of 0.99 (Fig. 2). The confusion matrix is presented in Fig. 3.
The class activation heatmaps (CAM) of OSCC and normal oral mucosae are illustrated in Figs. 4 and 5. These heatmaps visualize the discriminative regions used by the Swin-Transformer for the classification. Optical inspection indicates a more centered and focused region of interest for OSCC. For normal mucosa, either a blank heatmap without any focus or a widely distributed focus was noticed.
Discussion
Oral squamous cell carcinoma is a common malignancy with overall high mortality and morbidity2,3,4. The lack of experience and training of primary health care professionals leads to diagnostic delays and consequently to more extensive surgical procedures with more extended hospitalization and lower survival rates18,19,20. An automated assistance system for the clinician may increase the diagnostic accuracy while reducing the observer dependency. The vision transformer network introduced in this study is an accurate tool for the classification of OSCC based on clinical photographs that are most often acquired for documentation purposes.
In the past, diagnostic methods such as vital staining, autofluorescence and chemiluminescence, narrow band imaging, and optical spectroscopy have been introduced and documented with varying sensitivity and specificity21. Vital staining had a sensitivity of 92.3% but most studies did not report the specificity. For autofluorescence, heterogeneous values of 50–100% for sensitivity and 12.5 and 75.5% for specificity were reported. Narrow band imaging showed high sensitivity between 84.62 and 93.93%, and specificity between 75.7 and 94.56%.
Different studies have recently applied CNNs to classify oral cancer from an oral photograph. Warin et al. reported a F1-score of 0.9875 and an AUC of 0.99 with DenseNet12113. Fu et al. achieved a similar F1-score (0.935–0.995) with a two-step approach15. In the first step, a Single Shot MultiBox Detector was applied to detect the region of interest. Subsequently, DenseNet assessed the pre-selected region of interest in the presence of OSCC. Welikala et al. achieved a significantly lower F1-score of 0.8707 with ResNet-10122. Shamim et al. compared multiple CNNs (e.g. AlexNet, GoogleNet, Inceptionv3, ResNet50, SqueezeNet and VGG19) to classify tongue lesions and achieved F1-scores ranging from 0.9048 to 0.975623.
However, a direct comparison of these previous studies should be regarded with caution. The performance of the CNNs is highly dependent on the dataset, the hyperparameters and the architecture itself24. The number of training and test sets varied greatly in the previous studies, and the data representativeness was unclear. Furthermore, clinical photographs were not standardized, and a high discrepancy was expected in perspective. For these reasons, the replication and validation of the previous results remain impracticable.
In the current study, the Swin-Transformer achieved a F1-score of 0.98 and an AUC of 0.99. The model had one false positive prediction and one false negative prediction, independent of location, staging or grading. Two key concepts are essential for high performance: hierarchical feature maps and shifted window attention. Firstly, hierarchical feature maps allow the intermediate tensors to be merged from layer to layer, reducing the spatial dimension (i.e. downsampling) of the feature maps effectively. In comparison to CNNs, patch merging is applied for downsampling instead of convolution operations. Secondly, the Swin-Transformer replaced the standard multi-head self-attention with a window and shifted window self-attention. The standard multi-head self-attention performs a global self-attention, resulting in a quadratic complexity. For this reason, the window self-attention computes attention only locally within specified windows. The shifted window self-attention addresses global information loss using cross-window connections between different layers. These two modules reduce the quadratic complexity to linear complexity17.
Although a high performance was achieved using a transformer, there are limitations. The reported study is limited by its monocentric design resulting in a database consisting of the local population. The photographic images were acquired with high-quality cameras and did not regard clinical settings in which images may be acquired with cameras or mobile devices with lower image quality. The Swin-Transformer are strictly confined to the employed train- and test set and may perform worse in real-world scenarios. Prospective studies are required to evaluate the diagnostic accuracy of the Swin-Transformer in a clinical setting.
In conclusion, the Swin-Transformer forms a promising foundation for further developing automatic screening of OSCC on clinical photographs. Deep learning-based assistance of clinicians may raise the rate of early detection of oral cancer and hence the survival rate and quality of life of patients.
Data availability
The datasets analyzed in the current study are not publicly available due to data protection but are available from the corresponding author on reasonable request.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Ragin, C. C. R., Modugno, F. & Gollin, S. M. The epidemiology and risk factors of head and neck cancer: A focus on human papillomavirus. J. Dent. Res. 86(2), 104–114 (2007).
Messadi, D. V., Wilder-Smith, P. & Wolinsky, L. Improving oral cancer survival: the role of dental providers. J. Calif. Dent. Assoc. 37(11), 789 (2009).
Chinn, S. B. & Myers, J. N. Oral cavity carcinoma: Current management, controversies, and future directions. J. Clin. Oncol. 33(29), 3269 (2015).
Tanriver, G., Soluk Tekkesin, M. & Ergen, O. Automated detection and classification of oral lesions using deep learning to detect oral potentially malignant disorders. Cancers 13(11), 2766 (2021).
Varela-Centelles, P. et al. Key points and time intervals for early diagnosis in symptomatic oral cancer: A systematic review. Int. J. Oral Maxillofac. Surg. 46(1), 1–10 (2017).
Coca-Pelaz, A. et al. Head and neck cancer: A review of the impact of treatment delay on outcome. Adv. Ther. 35(2), 153–160 (2018).
Epstein, J. B., Güneri, P., Boyacioglu, H. & Abt, E. The limitations of the clinical oral examination in detecting dysplastic oral lesions and oral squamous cell carcinoma. J. Am. Dent. Assoc. 143(12), 1332–1342 (2012).
Seoane, J., Warnakulasuriya, S., Varela-Centelles, P., Esparza, G. & Dios, P. D. Oral cancer: Experiences and diagnostic abilities elicited by dentists in North-western Spain. Oral Dis. 12(5), 487–492 (2006).
Van der Waal, I. Are we able to reduce the mortality and morbidity of oral cancer; some considerations. Med. Oral Patol. Oral Cir. Bucal. 18(1), e33 (2013).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med Image Anal. 42, 60–88 (2017).
Matsoukas, C., Haslum, J. F., Söderberg, M., & Smith, K. Is it time to replace cnns with transformers for medical images? Preprint at https://arxiv.org/pdf/2108.09038.pdf (2021).
Warin, K., Limprasert, W., Suebnukarn, S., Jinaporntham, S. & Jantana, P. Performance of deep convolutional neural network for classification and detection of oral potentially malignant disorders in photographic images. Int. J. Oral Maxillofac. Surg. 51(5), 699–704 (2022).
Song, B. et al. Automatic classification of dual-modalilty, smartphone-based oral dysplasia and malignancy images using deep learning. Biomed. Opt Express 9(11), 5318–5329 (2018).
Fu, Q. et al. A deep learning algorithm for detection of oral cavity squamous cell carcinoma from photographic images: A retrospective study. EClinicalMedicine 27, 100558 (2020).
Vinayahalingam, S. et al. Classification of caries in third molars on panoramic radiographs using deep learning. Sci. Rep. 11(1), 12609 (2021).
Liu, Z. et al.. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 10012–10022 (2021).
Scully, C. & Kirby, J. Statement on mouth cancer diagnosis and prevention. Br. Dent. J. 216(1), 37–38 (2014).
Conley, B. A. Treatment of advanced head and neck cancer: what Lessons have we learned?. J. Clin. Oncol. 24(7), 1023–1025 (2006).
Neal, R. D. et al. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer 112(1), 92–107 (2015).
Mazur, M. et al. In vivo imaging-based techniques for early diagnosis of oral potentially malignant disorders—Systematic review and meta-analysis. Int. J. Environ. Res. 18(22), 11775 (2021).
Welikala, R. A. et al. Automated detection and classification of oral lesions using deep learning for early detection of oral cancer. IEEE Access 8, 132677–132693 (2020).
Shamim, M. Z., Syed, S., Shiblee, M., Usman, M. & Ali, S. Automated detection of oral pre-cancerous tongue lesions using deep learning for early diagnosis of oral cavity cancer. Comput. J. 65(1), 91–104 (2022).
Vinayahalingam, S. et al. Automated chart filing on panoramic radiographs using deep learning. J. Dent. 115, 103864 (2021).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
T.F. Contributed to conception and design, acquisition, analysis, and interpretation of data; drafted and critically revised the manuscript. R.G.: Contributed to analysis and interpretation and critically revised the manuscript. A.S.: Contributed to data acquisition, analysis, and interpretation, and critically revised the manuscript. D.T.: Contributed to analysis and interpretation and critically revised the manuscript. M.H.: Contributed to conception and design, drafted and critically revised the manuscript. N.v.N.: Contributed to statistical evaluation, interpretation of data, and critically revised the manuscript. S.V.: Contributed to conception, design, data analysis, statistical evaluation, and data interpretation and critically revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Flügge, T., Gaudin, R., Sabatakakis, A. et al. Detection of oral squamous cell carcinoma in clinical photographs using a vision transformer. Sci Rep 13, 2296 (2023). https://doi.org/10.1038/s41598-023-29204-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29204-9
This article is cited by
-
Deep learning in oral cancer- a systematic review
BMC Oral Health (2024)
-
Dysregulation of TCONS_00006091 contributes to the elevated risk of oral squamous cell carcinoma by upregulating SNAI1, IRS and HMGA2
Scientific Reports (2024)
-
Artificial intelligence for classification and detection of oral mucosa lesions on photographs: a systematic review and meta-analysis
Clinical Oral Investigations (2024)
-
Supremacy of attention-based transformer in oral cancer classification using histopathology images
International Journal of Data Science and Analytics (2024)
-
Speech disorders in patients with Tongue squamous cell carcinoma: A longitudinal observational study based on a questionnaire and acoustic analysis
BMC Oral Health (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.