Abstract
Predictive factors for response to regorafenib in recurrent glioblastoma, IDH-wildtype, are scarcely recognized. The objective of this study was to identify molecular predictive factors for response to regorafenib using a clinically available platform. We analyzed a prospective cohort of 30 patients harboring recurrent glioblastoma, IDH-wildtype, and treated with regorafenib. Next-generation sequencing (NGS) analysis was performed on DNA extracted from paraffin-embedded tissues using a clinically available platform. Moreover, MGMT methylation and EGFRvIII expression analyses were performed. Six-month progression-free survival (PFS) was 30% and median overall survival (OS) was 7.5 months, in line with literature data. NGS analysis revealed a mutation in the EGFR pathway in 18% of cases and a mutation in the mitogen-activated protein-kinase (MAPK) pathway in 18% of cases. In the remaining cases, no mutations were detected. Patients carrying MAPK pathway mutation had a poor response to regorafenib treatment, with a significantly shorter PFS and a nonsignificantly shorter OS compared to EGFR-mutated patients (for PFS, 2.5 vs 4.5 months, p = 0.0061; for OS, 7 vs 9 months, p = 0.1076). Multivariate analysis confirmed that MAPK pathway mutations independently predicted a shorter PFS after regorafenib treatment (p = 0.0188). The negative prognostic role of MAPK pathway alteration was reinforced when we combined EGFR-mutated with EGFRvIII-positive cases. Recurrent glioblastoma tumors with an alteration in MAPK pathway could belong to the mesenchymal subtype and respond poorly to regorafenib treatment, while EGFR-altered cases have a better response to regorafenib. We thus provide a molecular selection criterion easy to implement in the clinical practice.
Similar content being viewed by others
Introduction
Treatment of recurrent glioblastoma (GBM), IDH-wildtype, after surgery and radiotherapy with concomitant and adjuvant temozolomide remains a challenge. Failure of many trials with targeted drugs, including bevacizumab, led to the adoption of lomustine as standard second-line treatment in Europe1. The introduction of regorafenib changed this scenario. Regorafenib is an orally available multikinase inhibitor, whose main targets are kinases involved in angiogenesis (VEGFR1–3 and TIE2), oncogenesis (KIT, RET, RAF1, and BRAF), tumor microenvironment (PDGFR and FGFR), and tumor immunity (colony stimulating factor 1 receptor)2,3. In the REGOMA trial4, patients with recurrent GBM were randomized to receive regorafenib or lomustine. The regorafenib arm showed a significantly longer overall survival (OS), both in terms of median OS (7.4 vs 5.6 months, respectively; p = 0.0009) and 12-month OS (38.9% vs 15%, respectively). Safety was deemed acceptable. On these bases, regorafenib has become the standard second-line treatment for GBM, IDH-wildtype in our country. This notwithstanding, the setting of recurrence is particularly hard, and many issues remain unresolved. e.g., in the REGOMA trial, at 6 months from the start of regorafenib more than 80% of patients had experienced disease progression. It is therefore mandatory to identify those patients who are likely to respond poorly to regorafenib. Due to its novelty, available literature data are scarce5,6. The objective of this study was to analyze the molecular profile of a cohort of patients suffering from recurrent GBM, IDH-wildtype, treated with regorafenib, with the aim to identify factors that may predict response to this multikinase inhibitor.
Methods
Study outline
We prospectively enrolled patients harboring recurrent GBM, IDH-wildtype, after surgery and standard chemoradiotherapy according with the Stupp protocol7, and treated with regorafenib in the multidisciplinary Neuro-Oncologic Board at Fondazione Policlinico Universitario “A. Gemelli”, Rome, Italy. Indications and schedule of regorafenib treatment followed the executive decision of the Italian Agency for Medicines 143345/2019. Regorafenib was administered orally at a dose of 160 mg/day for the first 3 weeks of each 4-week cycle4. Neurosurgery for recurrent tumor before regorafenib treatment was allowed, when possible, to confirm the diagnosis of tumor recurrence. The study followed the principles set forth in the World Medical Association Declaration of Helsinki and was approved by the local Institutional Ethics Committee (Prot. ID 3458). Informed consent was obtained from all individual participants included in the study.
Standard histopathology and molecular profiling of the tumor
Enrolled patients were diagnosed with GBM, IDH-wildtype, according to WHO classification of tumours of the Central Nervous System, revised 4th edition8. The presence of IDH1 R132H mutation was assessed by immunohistochemistry using the monoclonal antibody clone H09 (Dianova, Hamburg, Germany). MGMT promoter methylation analysis was performed with methylation-specific PCR, and EGFRvIII expression was assessed by RT-PCR, as previously described9.
Analysis of next-generation sequencing data for sequence variants and copy number changes
Next-generation sequencing (NGS) analysis was performed on DNA extracted from paraffin-embedded tissues with a percentage of tumor cells higher than 50%, using the QIAcube system (Qiagen, Hilden, Germany). The DNA was then amplified using Myriapod NGS Cancer panel DNA, REF NG033, Kit IVD (Diatech Pharmacogenetics, Jesi, Italy) on EasyPGX qPCR instrument 96 (Diatech Pharmacogenetics), and the sequencing was done using Seq100 (Illumina, San Diego, CA). The sensitivity of the procedure was around 5%. The analyzed genes were ALK, BRAF, EGFR, ERDD2, FGFR3, HRAS, IDH1, IDH2, KIT, KRAS, MET, NRAS, PDGFR, PIK3CA, RET, ROS1. Data analysis was performed using Myriapod NGS Data Analysis Software and Myriapod NGS Workstation (Diatech Pharmacogenetics).
Statistical analysis
Response to treatment was evaluated using Response Assessment in Neuro-Oncology (RANO) criteria10. Progression-free survival (PFS) and OS were calculated from regorafenib start to, respectively, progression of disease and patient’s death. The primary endpoint of the study was the correlation between molecular profile and PFS after regorafenib treatment. Secondary endpoints were the correlation between molecular profile and OS and response to treatment. Comparison of categorical variables was conducted with the Chi-square statistics, using the Fisher Exact test when appropriate. Comparison of continuous variables between groups was conducted using the Mann–Whitney U test. For survival analysis, Kaplan–Meier curves were plotted and comparison between groups was performed using the log-rank test. In the whole series, multivariate analysis of prognostic factors for PFS and OS was performed by using the Cox model, adjusted for the following parameters, age (< 65 years or ≥ 65 years), extent of resection, MGMT promoter methylation, EGFRvIII status, NGS analysis. Another multivariate analysis of prognostic factors for PFS and OS in patients with MAPK or EGFR pathway alteration was performed by using the Cox model, adjusting for the following parameters: age (< 65 years or ≥ 65 years), extent of resection, MGMT promoter methylation, molecular status. Median follow-up was calculated using the reverse Kaplan–Meier method. A p value less than 0.05 was considered as statistically significant. Analyses were performed using StatView ver 5.0 (SAS Institute, Cary, NC).
Ethics approval
The study followed the principles set forth in the World Medical Association Declaration of Helsinki and was approved by the Institutional Ethics Committee of Fondazione Policlinico Gemelli IRCCS (Prot. ID 3458). We confirm that informed consent was obtained from all from all subjects (and/or their legal guardian(s)) included in the study.
Results
Baseline patients characteristics
Thirty recurrent GBM patients after standard radiotherapy with concomitant and adjuvant chemotherapy according to the Stupp protocol were considered in this study. NGS analysis showed that one of the patients harbored IDH-mutant GBM that was not detected using immunohistochemistry. Therefore, this patient was excluded from the study. Then, the final group was composed of 29 patients harboring GBM, IDH-wildtype (Online Resource 1, Supplementary Table S1).
At enrollment, the mean age was 58.4 ± 10.8 years. All patients had a good performance status (i.e., Karnofsky performance status ≥ 70). Fourteen patients (48.3%) had undergone surgery for recurrent GBM. At last surgery, gross-total tumor resection had been performed in 22 out of 29 cases (75.9%), whereas subtotal resection or biopsy in 7/29 cases (24.1%). Among patient who underwent surgery for recurrence, gross-total tumor resection was performed in 10 out of 14 cases (71.4%), whereas subtotal resection in 4/14 cases (28.6%). MGMT promoter was methylated in 12 out of 29 cases (41.4%) and unmethylated in 17/29 (58.6%). RT-PCR for EGFRvIII mRNA was available for 28 patients and was positive in 14 cases.
Treatment results
Median follow-up was 20 months (range, 1.5–20). Dose reduction was required in 8 patients; reasons were thrombocytopenia (2 cases), fatigue (2 cases), hand-foot syndrome (2 cases), diarrhoea (1 case), and hyperbilirubinemia (1 case). At last follow-up, 8 out of 29 patients (27.5%) were alive, two of them with ongoing regorafenib treatment while the other six patients experienced disease progression during regorafenib treatment; 21/29 patients (72.5%) were dead. The median PFS of the whole cohort was 3 months. Six-month PFS (PFS-6) with regorafenib treatment was reached in 8 out of 27 patients (29.6%) and was not reached in 19/27 patients (70.4%). Median OS was 7.5 months. The best response was assessed using RANO criteria10, in which partial response (PR) requires ≥ 50% reduction of contrast enhancement without increase in FLAIR alterations, progressive disease (PD) requires a ≥ 25% increase of contrast enhancement or increase in FLAIR alterations or any new contrast enhancing lesion, and stable disease (SD) does not qualify either for PR or for PD. Excluding the 2 cases with ongoing regorafenib treatment, the best response was partial response (PR) in 5 cases (18.5%), stable disease (SD) in 8 cases (29.6%), and progressive disease (PD) in 7 cases (25.9%). In 7 other cases (25.9%), PD was diagnosed on clinical basis only.
Next-generation sequencing analysis
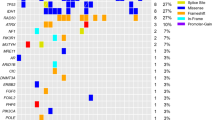
NGS analysis was successfully conducted in 28 out of 29 cases. In 1 case, extracted DNA was not sufficient. In 5 out of 28 cases (17.9%), mutation of EGFR and/or PIK3CA was found. These cases were grouped together, as cases harboring mutation in the EGFR pathway. RAS mutations were found in two cases (7.1%) and RET mutations were found in 3 cases (10.1%). These cases were grouped together as cases harboring mutation in the mitogen-activated protein kinase (MAPK) pathway. In the remaining 18 cases (64.3%), no mutations of the analyzed genes were found. Details on the specific mutations are given in Online Resource 1, Supplementary Table S2.
Standard prognostic factors for progression-free and overall survival after regorafenib treatment
Age had no prognostic value, probably due to the careful patients’ selection (Online Resource 2, Supplementary Fig. S1). Gross-total resection (GTR), instead, was endowed with a positive prognostic value for PFS (median PFS 4 months in GTR cases vs 2.5 months in subtotal resection/biopsy cases, p = 0.0296, log-rank test) but not for OS (Online Resource 2, Supplementary Fig. S1). Patients whose tumors had a methylated MGMT promoter had a median PFS of 4 months, while in patients with unmethylated MGMT promoter median PFS was 3 months, however, this difference was not significant (p = 0.42, log-rank test) (Online Resource 2, Supplementary Fig. S1). MGMT promoter methylation did not impact on OS (median OS 7.5 months both in methylated and unmethylated cases, p = 0.13, log-rank test). EGFRvIII expression conferred a not significantly longer PFS (median PFS 4 months in EGFRvIII positive vs 2.5 months in EGFRvIII negative cases, p = 0.40, log-rank test) and a significantly longer OS after regorafenib treatment (median PFS 8.5 months in positive vs 4.5 months in negative cases, p = 0.0131, log-rank test) (Online Resource 2, Supplementary Fig. S1).
Prognostic role of next-generation sequencing analysis after Regorafenib treatment
Based on the analyzed genes, we categorized patients into three groups, those carrying a mutation in the EGFR pathway (n = 5), those carrying a mutation in the MAPK pathway (n = 5), and those with no detectable mutations (n = 18). Surprisingly, patients carrying a MAPK pathway mutation had a poor response to regorafenib treatment, with median PFS 2.5 months, which was significantly shorter compared to patients carrying EGFR pathway mutation (4.5 months) and wild-type cases (3.5 months) (p = 0.0480; p = 0.0061 for comparison between MAPK and with EGFR mutated patients; log-rank test) (Fig. 1). Overall survival analysis confirmed that patients harboring MAPK pathway mutation had shorter OS than those with EGFR pathway mutation, but the difference was not significant (median OS 7 vs 9 months, respectively; p = 0.1076; log-rank test).
Multivariate Cox analysis for PFS confirmed the independent negative predictive role of MAPK pathway alteration for response to regorafenib treatment (p = 0.0188; Table 1), while no independent prognostic factors for OS were identified (Online Resource 1, Supplementary Table S3).
Prognostic role of combination of NGS-EGFRvIII analysis after regorafenib treatment
In 18 out of 28 cases (64%), NGS analysis did not identify any gene mutation; however, 9 of them (50%) expressed EGFRvIII as showed by RT-PCR analysis, portending an EGFR pathway activation. Therefore, we grouped these 9 patients together with the 5 cases harboring activating mutation of the EGFR pathway (n = 14 overall), and compared PFS and OS of this group with survival of patients harboring MAPK pathway mutation (n = 5). As shown in Fig. 2, patients with MAPK pathway alteration had significantly worse PFS and OS than those with an EGFR pathway alteration (median PFS 2.5 months vs 4 months, respectively; p = 0.0004; median OS 7 vs 9 months, respectively; p = 0.0242; log-rank test). Multivariate analysis confirmed the independent negative prognostic role of MAPK pathway activation compared to EGFR pathway activation, both for PFS (p = 0.0034; Table 2) and OS (p = 0.0426; Online Resource 1, Supplementary Table S4).
Discussion
Glioblastoma recurrence is one of the main challenges in neuro-oncology. Various salvage strategies have been applied with limited success. The randomized multicenter Phase II REGOMA trial showed that regorafenib significantly improved OS when compared to lomustine (7.4 vs 5.6 months)4. Considering the extreme heterogeneity of this tumor, we performed NGS analysis of a panel of genes in patients suffering from recurrent GBM, IDH-wildtype treated with regorafenib, to identify predictive factors for response to treatment. Few studies in the literature are available on this topic5,6. Remarkably, we observed a median PFS of 3 months and a median OS of 7.5 months, comparable to those reported in the REGOMA trial4.
The main result of our study is that patients harboring an alteration in the MAPK pathway, as assessed by NGS at DNA level, display poor response to regorafenib in terms of PFS, which was the primary endpoint of the study. Combining NGS analysis with RT-PCR for EGFRvIII, MAPK-altered patients had a significantly worse prognosis than EGFR-altered patients in terms of OS.
Standard clinical and molecular prognostic factors of response to regorafenib
Noteworthy, all patients had KPS ≥ 70 at enrollment; it is thus not surprising that age was not a prognostic factor in this group, differently from unselected GBM cohorts. According with recent literature11,12, gross-total resection positively impacted on the clinical outcome. No patient undergoing subtotal resection or biopsy reached 6-month PFS. When considering separately patients who underwent surgery for recurrence, a trend to improved PFS with gross-total resection was confirmed at log-rank analysis (p = 0.0809).
We found higher PFS-6 rate in MGMT methylated than unmethylated patients, though PFS advantage at Kaplan–Meier analysis was not significant. This evidence has not an obvious mechanistic explanation. In the REGOMA trial, survival advantage with regorafenib was noticed both in unmethylated and methylated tumors with a greater amplitude in the latter4. Intriguingly, EGFRvIII mRNA expression conferred a nonsignificantly longer PFS and a significantly longer OS after regorafenib treatment. This evidence also is puzzling, since regorafenib is not an EGFR inhibitor. In a previous work from our group on a cohort of 73 primary GBM patients9, EGFRvIII-positive patients had median OS of 19 months, which was significantly longer than EGFRvIII negative ones. Thus, we could argue that the prognostic advantage of EGFRvIII-positive patients was independent from regorafenib treatment. In the present series, however, the cumulative OS, as calculated since GBM diagnosis, was not significantly different between EGFRvIII-positive and negative cases (22.5 vs 22 months, respectively; p = 0.7; log-rank test). It should be underlined that the work by Montano et al9 included a large cohort of unselected patients, whereas in the present study the patients were selected for their clinical conditions (KPS ≥ 70). Therefore, the EGFRvIII expression was a definite predictor of good response to regorafenib.
Advanced molecular prognosticators of response to regorafenib
Regorafenib is a multikinase inhibitor, whose main targets are the VEGF receptors, the members of the ERK-MAPK pathways (RET, RAF-1, BRAF) and other kinases like KIT, PDGFR and FGFR. Due to its recent introduction into neuro-oncology practice, molecular factors for response are poorly known. The Padua group, which led the REGOMA trial, published two papers where, (i) the immunohistochemical expression of phosphorylated acetyl-CoA carboxylase(pACC)6, and (ii) the mini-signature including HIF1A and CDKN1A mRNAs, besides 3 miRNAs, showed positive predictive value for response to regorafenib5. Mechanistically, HIF1A, CDKN1A, and the 3 miRNAs are angiogenesis players: their positive predictive value for response to regorafenib is therefore explained by the anti-angiogenic activity of regorafenib. Antiangiogenic treatment would trigger the 5’adenosine monophosphate-activated protein kinase (AMPK) and prime metabolic rewiring, which restrains cell proliferation. Interrogating ACC phosphorylation in tumors can indicate whether the pathway is active. Therefore, pACC-positive tumors are thought to be more angiogenic compared with pACC-negative ones. However, this hypothesis was not supported by microvasculature density analysis, which suggested similar angiogenesis in pACC-positive and pACC-negative tumors. In the pAMPK-positive cohort of patients treated with regorafenib, there was improved OS, but this biomarker yielded negative results in the interaction test.
In our analysis, we identified mutations in 36% of GBM tumors, which involved the EGFR, PIK3CA, Ras and RET genes (Online Resource 3, Supplementary Discussion). EGFR/PI3KA mutations are one of the most frequent events in gliomagenesis13. Ras is a known player in the ERK-MAPK pathways14. RET is a receptor tyrosine kinase, whose dimerization and subsequent internalization activates a variety of downstream pathways, including the MAPK/ERK pathway14. Reportedly, the RET mutations that were found in this study activate specifically the MAPK pathway (Online Resource 3, Supplementary Discussion)15. Then, it is somewhat counter-intuitive that activating mutations of this pathway, which is a known target of regorafenib, are predictive of poor response. While the scarcity of available data hampers a definitive explanation, some speculations can be made. We might postulate that the mutations we found render the tyrosine kinases RET and RAF (the downstream effector of Ras) resistant to regorafenib inhibition16,17. Another intriguing explanation concerns the ERK-MAPK pathway, which is regarded as the core pathway of mesenchymal GBM18. It is not surprising that regorafenib, an anti-angiogenic drug, has a scarce activity on mesenchymal GBM. Bevacizumab, another anti-angiogenic drug, is reportedly more effective on proneural GBMs19. The mesenchymal subtype of GBM displays higher metabolic flexibility with both glycolytic and oxidative metabolisms. In contrast to proneural ones, the mesenchymal GBMs easily bypass targeted metabolic inhibition through a wide range of metabolic adaptation. To overcome their metabolic flexibility, an efficient metabolic targeting of mesenchymal GBM will require the blockade of several metabolic pathways. For example, in preclinical cancer models, dual inhibition of both glycolysis and oxidative phosphorylation has been shown to effectively reduce tumor growth20. To conclude, the NGS panel used in this analysis could identify a subgroup of tumors with activated MAPK pathway, putatively mesenchymal recurrent GBMs, which responded poorly to regorafenib.
Strength of the present study
We present a series of patients homogeneously treated at a nationwide neuro-oncologic referral center, where clinical decisions and follow-up are regularly made by the multidisciplinary neuro-oncologic team. The prospective enrollment of patient assures the high quality of data. Another strong point of this work is linked to the NGS platform (Illumina ISeq™ 100). This is a rapid system, which does not require demanding hardware, and thanks to CE/IVD validation can be used for diagnostic purpose. The system allows the sequencing of small groups of patients per run (8–10 patients) and has reduced costs (about 150 euro per patient). Therefore, it allows time-sensitive genomic analysis of patients candidate to chemotherapy.
Limitations of the study
The monocentric nature of the study can be regarded as a limitation, though, as stated before, it assures the quality of clinical and molecular data. The sample size is relatively small, and this can impact on statistical significance. In this study, we used PFS as primary endpoint, while most studies on recurrent GBM use as endpoint the OS, in order to overcome uncertainties in assessing progression4,21. However, we believe that PFS is an adequate endpoint for the assessment of predictive factors for response to regorafenib, as it is not influenced by the responses to first-line treatment or to third-line therapies.
Conclusions
Our data suggest that NGS analysis using a simple and cheap platform, combined with standard molecular profiling, might be used in patients candidate to second-line chemotherapy in order to select patients who can benefit from regorafenib treatment. Such approach could be useful in our clinical practice to customize the second-line strategy, taking into account patient’s comorbidities and the possible adverse events related to different treatments. Although far from being conclusive, these data support the development of further studies.
Data availability
The datasets analysed during the current study are available in the clinVAR repository (www.ncbi.nlm.nih.gov/clinvar/). The 3 variants not available in clinVAR repository have already been described (See Online Resource 1, Supplementary Table S2).
Abbreviations
- EGFR:
-
Epidermal growth factor receptor
- EGFRvIII:
-
Epidermal growth factor receptor variant III
- GBM:
-
Glioblastoma
- GTR:
-
Gross-total resection
- MAPK:
-
Mitogen-activated protein-kinase
- NGS:
-
Next-generation sequencing
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PFS-6:
-
Six-month progression-free survival
- PD:
-
Progressive disease
- PR:
-
Partial response
- RANO:
-
Response Assessment in Neuro-Oncology
- SD:
-
Stable disease
References
Weller, M. & Le Rhun, E. How did lomustine become standard of care in recurrent glioblastoma?. Cancer Treat. Rev. 87, 102029 (2020).
Wilson, T. A., Karajannis, M. A. & Harter, D. H. Glioblastoma multiforme: State of the art and future therapeutics. Surg. Neurol. Int. 5, 64 (2014).
Wilhelm, S. M. et al. Regorafenib (BAY 73–4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 129, 245–255 (2011).
Lombardi, G. et al. Regorafenib compared with lomustine in patients with relapsed glioblastoma (REGOMA): A multicentre, open-label, randomised, controlled, phase 2 trial. Lancet Oncol. 20, 110–119 (2019).
Santangelo, A. et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 23, 264–276 (2020).
Indraccolo, S. et al. Phosphorylated acetyl-CoA carboxylase is associated with clinical benefit with regorafenib in relapsed glioblastoma: REGOMA trial biomarker analysis. Clin. Cancer Res. 26, 4478–4484 (2020).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 131, 803–820 (2016).
Montano, N. et al. Expression of EGFRvIII in glioblastoma: Prognostic significance revisited. Neoplasia 13, 1113–1121 (2011).
Wen, P. Y. et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J. Clin. Oncol. 28, 1963–1972 (2010).
Lauretti, L. et al. Molecular analysis in a glioblastoma cohort-results of a prospective analysis. J. Pers. Med. 12, 685 (2022).
Della Pepa, G. M. et al. 5-Aminolevulinic acid and contrast-enhanced ultrasound: The combination of the two techniques to optimize the extent of resection in glioblastoma surgery. Neurosurgery 86, E529–E540 (2020).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Richardson, D. S., Lai, A. Z. & Mulligan, L. M. RET ligand-induced internalization and its consequences for downstream signaling. Oncogene 25, 3206–3211 (2006).
Sawai, H. et al. The G691S RET polymorphism increases glial cell line-derived neurotrophic factor-induced pancreatic cancer cell invasion by amplifying mitogen-activated protein kinase signaling. Cancer Res. 65, 11536–11544 (2005).
Hamed, H. A., Tavallai, S., Grant, S., Poklepovic, A. & Dent, P. Sorafenib/regorafenib and lapatinib interact to kill CNS tumor cells. J. Cell Physiol. 230, 131–139 (2015).
Bauer, S., George, S., von Mehren, M. & Heinrich, M. C. Early and next-generation KIT/PDGFRA kinase inhibitors and the future of treatment for advanced gastrointestinal stromal tumor. Front. Oncol. 11, 672500 (2021).
Park, A. K., Kim, P., Ballester, L. Y., Esquenazi, Y. & Zhao, Z. Subtype-specific signaling pathways and genomic aberrations associated with prognosis of glioblastoma. Neuro Oncol. 21, 59–70 (2019).
Sandmann, T. et al. Patients with proneural glioblastoma may derive overall survival benefit from the addition of bevacizumab to first-line radiotherapy and temozolomide: Retrospective analysis of the AVAglio trial. J. Clin. Oncol. 33(2735), 2744 (2015).
Chen, C. et al. Osimertinib successfully combats EGFR-negative glioblastoma cells by inhibiting the MAPK pathway. Acta Pharmacol. Sin. 42, 108–114 (2021).
Wick, W. et al. Lomustine and bevacizumab in progressive glioblastoma. N. Engl. J. Med. 377, 1954–1963 (2017).
Funding
The work was supported by Associazione Italiana per la Ricerca sul Cancro (IG 2019 23154 to R.P.).
Author information
Authors and Affiliations
Contributions
Conception: R.P., M.M., M.B. Design: R.P., M.M., M.B., S.C., Q.G.D., Acquisition of data: all Authors. Analysis, or interpretation of data: all Authors. Statistical analysis: QGD. Drafted the work: R.P., M.M., M.B. S.C., Q.G.D., A.M. Revised it critically for important intellectual content: all Authors. Approved the version to be published: all Authors. Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: all Authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chiesa, S., Mangraviti, A., Martini, M. et al. Clinical and NGS predictors of response to regorafenib in recurrent glioblastoma. Sci Rep 12, 16265 (2022). https://doi.org/10.1038/s41598-022-20417-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20417-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.