Abstract
NPY-family receptors belong to G protein-coupled receptors (GPCR), which lays a physiological foundation for the transmembrane transport of an endogenous appetite-stimulating factor neuropeptide Y and related peptides. In this study, we investigated the npyr genes in ten representative species, and twelve npyr genes were identified from allotetraploid C. carpio, the npyr gene number of C. carpio was twice the number of its subgenome B progenitor-like diploid Poropuntius huangchuchieni. Phylogenetic analysis showed that all npyr genes were divided into three subgroups, and they underwent strong purifying selection according to selection pressure analysis. Subsequently, synteny analysis showed that most npyr genes were evenly distributed on the homologous chromosomes of two subgenomes in allotetraploid C. carpio, in which npy1r and npy2r were tandem duplicated, respectively. In addition, the global expression of npyr genes during embryonic development in allotetraploid C. carpio suggested the potential function of npyr genes in immunity and reproduction. In adult tissues, npyr genes were mainly distributed in the brain, gonad, and skin, which displayed a similar expression pattern between the C. carpio B subgenome and P. huangchuchieni. In general, our research could provide reference information for future exploration of the NPY receptors and neuroendocrine system of allotetraploid C. carpio and vertebrates.
Similar content being viewed by others
Introduction
Neuropeptide Y (NPY) and its related peptides usually affect the neuroendocrine system of organisms by acting on several G protein-coupled receptors1, including the regulation of appetite2 and response to anxiety3 as well as surrounding pressure4, and act as immune roles5. NPY-family receptors are thought to be mainly expressed in fish brains, as well as in marginal tissues such as eyes and intestines6. Based on the structural characteristics of the receptor and the amino acid sequence similarity, NPY-family receptors are generally divided into seven subtypes: Y1, Y2, Y4, Y5, Y6, Y7 and Y86. They all belong to the G protein-coupled receptors. Among them, Y1, Y4, Y6, Y8a and Y8b belong to the subfamily of Y1 NPY-family receptor genes7. The fifth subtype Y6 usually exists in primates in the form of pseudogenes8. There is only one subtype in the Y5 category and it does not have an additional copy. Y2 and Y7 both belong to the Y2 subtype, which is due to the increased copy of gene replication9,10. All members of the NPY-family receptor are thought to have originated from a single receptor gene in the ancestors of all vertebrates through multiple rounds of whole-genome duplication8.
It has been reported that Y1, Y2, Y4, Y7, Y8a and Y8b are all present in teleost9. The Y1-like and Y5 receptor genes were found in basal teleost1,11, while Y1 and Y5 were thought to be missing in both zebrafish and pufferfish10,12. The loss of these genes may be a consequence of large-scale duplication in the process of lineage evolution10. But the Y1 receptor was identified in whole-genome data of zebrafish later13. Y1 receptors are considered to be receptors for promoting food intake, and studies in goldfish have shown that the effect of appetite-stimulating factors on food intake is partly mediated by Y1 receptors rather than Y2 receptors14. The Y2 receptor has attracted much attention as an appetite suppressor receptor expressed in the hypothalamic arcuate nucleus15, and the Y7 receptor is considered to be a direct homologue of Y28. The pharmacological properties of Y7 were studied in zebrafish, and it was found that the Y7 receptor was more sensitive to the truncation of amino terminals of peptide ligands than Y216. Both Yb and Yc, which were first identified in zebrafish, were identified as Y8a and Y8b later, and the two receptor genes were also identified in pufferfish. Their chromosome location showed that they were most likely to occur in teleost whole-genome duplication (3R)10,17.
The evolutionary dynamics of NPY and its receptors in whole-genome duplication events in early vertebrates have been thoroughly studied1,8. Similarly, the fate of NPY and its receptor genes in the third round of teleost-specific whole-genome duplication events (Ts3R WGD) have been studied thoroughly13,17. The NPY-family receptors of autotetraploid fish like salmon and trout have been identified, but the duplicated Y2 and Y7 receptors may not be found due to the incompleteness of the genomic DNA library12. The occurrence of autotetraploidy events in salmon and trout is relatively early18 and rediploidization and gene loss may have happened19. The evolutionary fate of NPY-family receptor genes in common carp that experienced the most recent polyploidy is still unknown.
As a type of polyploid fish widely distributed worldwide, common carp have strong stress resistance20,21,22. According to the Ka/Ks analysis of homologous genes and the divergence rate of transposable elements, it is inferred that the tetraploidy event of common carp may have occurred 12.4 million years ago (Mya) and coincided with the climate change event driven by the sharp uplift in the eastern Qinghai-Xizang Plateau23. The allotetraploid common carp can be divided into two sets of subgenome A and B according to the coverage of each of the 50 pairs of chromosomes between the related species23. Based on systematic genomic evidence, it has been determined that Poropuntius–Puntius–Hampala is a closely related diploid progenitor-like group of common carp B subgenome23, and representative diploid Poropuntius huangchuchieni and Onychostoma macrolepis were included in this study.
This study led to a better understanding of the evolutionary fate of npyr genes in allotetraploid common carp after the fourth round of whole-genome duplication events. The abundance and phylogeny relationships of npyr genes were analyzed in ten species, as well as allotetraploid common carp and related diploid and autotetraploid species. Structure and conservative motif analysis provide evidence for the classification of npyr genes. Based on the results above, we also conducted a comparative expression analysis of npyr genes in allotetraploid common carp and related diploid species. The abundance, distribution and expression profile of npyr genes in allotetraploid common carp provide us with more comprehensive information about the evolutionary footprint and functional exploration of npyr genes after the fourth round of whole-genome duplication event.
Materials and methods
Identification and nomenclature of npyr genes
In order to identify the npyr genes in the whole-genome of allotetraploid C. carpio23 and its subgenome B progenitor-like diploid P. huangchuchieni24, we used the sequences of npyr genes such as Y1, Y2, Y4, Y7and Y8a in Danio rerio, which were downloaded from Ensembl (https://uswest.ensembl.org/index.html) as queries. Both BLASTP and tBLASTn were used to search against newly published genomes for comprehensive investigations of npyr genes. The other seven species investigated in this study include Homo sapiens, diploid fish Oryzias latipes, Poropuntius huangchuchieni, Onychostoma macrolepis, Takifugu rubripes and tetraploid fishes Carassius auratus, Salmon salar and Oncorhynchus mykiss. The acronym was used to replace the Latin name of the species. For example, Homo sapiens was abbreviated as Hsa or H. sapiens. All npyr genes in C. carpio and other species were renamed according to its D. rerio ortholog (npy1r and npy2r, etc.), with species abbreviated as a prefix and numbered suffixes (Cca_npy1r-1, Cca_npy1r-2, etc.).
Phylogenetic analysis
Multiple sequence alignment was conducted by clustalx2.125 and visualized by ESPript (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi)26. All the NPY receptor sequences of ten species were aligned via MEGA X27 built-in alignment button MUSCLE28 and then manually trimmed. We performed the phylogenetic analysis with RA × ML (version 8.2.12) and obtained the maximum likelihood (ML) tree. Subsequently, the online tool EvoView (https://evolgenius.info/evolview-v2/#login)29 was used to visualize the phylogenetic tree, the same method was used in producing species trees based on their shared npy2r genes.
Structure and conservative motif analysis
We used a simple modular architecture research tool (SMART http://smart.embl-heidelberg.de/) to predict conserved domain architectures of NPY receptors in C. carpio. The results were visualized by IBS (version 1.0). In addition, motif analysis was performed by using MEME (https://meme-suite.org/meme/tools/meme)30, including the motif logo specific to different subtypes of NPY receptors were visualized through TBtools (v1.0695)31. Additionally, the protein structure of NPY receptors in C. carpio was predicted and represented by Protter32.
Calculation of selective pressure
We used the evolutionary ratio of non-synonymous substitutions (dN) and synonymous substitutions (dS)33 on the NPY-family receptor gene sites to characterize the selection pressure. The complete amino acid sequences of Y1, Y2, Y4, Y7, Y8a and Y8b receptors were aligned with the MUSCLE in MEGA X to eliminate terminators. Then we calculated the selection pressure of each site at the codon level through the Single Likelihood Ancestor Count (SLAC) method on the Datamonkey server (http://www.datamonkey.org/)34.
Synteny analysis and comparative expression profiles
To exhibit the syntenic relationship of the npyr genes in allotetraploid C. carpio and related diploid fishes, the syntenic plots were constructed with a multiple collinearity scan toolkit (MCScanX)35.
In order to compare the expression profiles of npyr genes in different tissues and embryo developmental stages in allotetraploid C. carpio and C. carpio B subgenome progenitor-like diploid P. huangchuchieni, we analyzed transcriptome datasets of 11 embryo developmental stages (Sperm, Egg, Zygote, Morula, Blastula, Gastrula, Neurula, Optic vesicle, Tail bud, Muscle contraction, 1dph) of C. carpio and 12 adult tissues of C. carpio and P. huangchuchieni using Hisat236 and Stringtie37, all of these tissues were taken from the healthy and untreated fish and the TPM (Transcripts per million) value was used to qualify gene expression levels of NPY receptor coding genes. The transcriptome dataset of C. carpio was deposited in NCBI, with accession number PRJNA689982 (https://dataview.ncbi.nlm.nih.gov/object/), and the National Genomics Data Center under accession number PRJCA004216 (https://bigd.big.ac.cn/), and the transcriptome dataset of P. huangchuchieni was available at the National Genomics Center under Bioproject number PRJCA002855 (https://bigd.big.ac.cn/gsub/submit/bioproject/subPRO004206/overview). The visualization was presented by the dynamic heatmap module of an online heatmap tool (http://www.omicshare.com/tools/Home/Soft/heatmap).
Ethics approval
Animal treatments in the study were conducted following the regulations of the Guide for Care and Use of Laboratory Animals and approved by the Committee of Laboratory Animal Experimentation at College of Ocean and Earth Sciences, Xiamen University.
Results
Genomic investigation of the npyr gene repertoire
A total of 12 npyr genes were identified in the common carp genome, including Y1, Y2, Y4, Y7, Y8a and Y8b, all of which have two copies, and were evenly distributed on the homologous chromosomes of two subgenomes (Table 1). The results showed that the length of the sequence of npyr genes varied from 370 to 381 amino acids (aa). Except for npy1r-1 and npy1r-2 which had two exons, all of the npyr genes in the common carp only had a single exon.
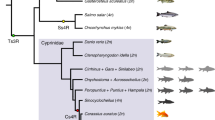
The number of npyr genes in all investigated species is shown in Fig. 1a. The results showed that the number of npyr genes of allotetraploid common carp was twice as much as that in related diploid species, such as D. rerio, O. macrolepis and P. huangchuchieni, which indicated that the common carp had undergone the fourth round of whole-genome duplication event and that the npyr genes were duplicated in a whole-genome duplication event. However, the related allotetraploid goldfish Y1 receptor had three copies, whereas Y4 and Y7 had only one copy (Fig. 1a). This meant that the npyr genes in common carp and goldfish had a different fate after the fourth round of whole-genome duplication events.
Comparison of gene copy numbers of npyr genes among selected vertebrate genomes from marine and freshwater habitats. (a) The phylogenetic tree of the species examined in this study was constructed using common Y2 receptor amino acid sequences. The blue rectangles represent marine fish, while the green ones represent freshwater fish. The colorful rectangles in the right panel are marked with the copy numbers of the npyr gene. (b) Amino acid sequence alignments of NPY receptors for all fish species examined. The conserved regions are indicated by a shadow. The black shadow indicates the region where all sequences share the same amino acid residue. The red bars indicate positions that vary between marine fish and freshwater fish. Species abbreviations include H. sapiens, Homo sapiens, D. rerio, Danio rerio, S. salar, Salmo salar, O. mykiss, Oncorhynchus mykiss, O. latipes, Oryzias latipes, P. huangchuchieni, Poropuntius huangchuchieni, O. macrolepis, Onychostoma macrolepis, C. carpio, Cyprinus carpio, T. rubripes, Takifugu rubripes and C. auratus, Carassius auratus.
As for autotetraploid, we found that the Y1 and Y4 receptors of D. rerio lacked orthologues in S. salar and O. mykiss, which have two copies of Y2, Y7, Y8a and Y8b. This showed that the doubling of npyr genes is indeed accompanied by a whole-genome duplication event. Regarding the loss of Y1 and Y4 receptors in two autotetraploids, we also searched for them in O. latipes and T. rubripes, and found that all marine fish (Fig. 1a) lack Y1 receptors compared with freshwater fish. The Y4 receptors are still retained in diploid species, but are missing in autotetraploids in marine habitats, which may be closely related to the early rediploidization event of autotetraploid19.
In order to confirm the characteristics of the npyr genes in teleosts from different habitats, we carried out multiple alignment of the npy2r amino acid sequences shared by teleosts in different habitats. All the sequence similarities were ranging from 70.25 to 95.92%, and the amino acid codes for pairs of conserved regions remained consistent with species of the same habitat, but there were exclusive site differences between different habitats. It was found that 12 residue sites were completely different between marine and freshwater habitats, for example, Y76F meant a mutation from Y to F at residue positions 76 among freshwater and marine habitats (Fig. 1b). We implied that these teleosts from two different habitats may have such possible species-specific mutation sites in completely conservative regions of the genome. These sites may be the main regulatory regions for the adaptive evolution of different habitats.
Phylogenetic analysis of NPY-family receptor genes
To investigate the evolutionary relationship between the npyr genes in vertebrate lineages, we constructed a phylogenetic tree using 71 amino acid sequences of npyr genes in ten investigated species (Fig. 2). All npyr genes were divided into three subgroups, with Y1, Y4, Y8a and Y8b belonging to the Y1 subgroup that was clustered into one group, and Y2 and Y7 belonging to the Y2 subfamily were clustered into the other group. While NPY5R in humans was a separate group, it had no orthologs in fish (Fig. 2).
Phylogenetic tree of the NPY-family receptors in vertebrates. All of the same types of NPY-family receptors are shown to have the same color backgrounds as the NPY-family receptors of common carp that are marked by red stars. The one marked with a triangle is NPY5R, which is used as an outgroup of the phylogenetic tree. Abbreviations for species names: Homo sapiens (Hsa), Danio rerio (Dre), Salmo salar (Ssa), Oncorhynchus mykiss(Omy), Oryzias latipes (Ola), Poropuntius huangchuchieni (Phu), Onychostoma macrolepis (Oma), Cyprinus carpio (Cca), Takifugu rubripes(Tru), Carassius auratus (Cau).
Selection pressure of teleost npyr genes (dN/dS)
To obtain the evolutionary dynamics of npyr genes in the vertebrate evolutionary lineage, we conducted a selective pressure analysis and calculated the substitution rates of non-synonymous substitutions (dN) and synonymous substitutions (dS) (Table 2). The ratio of dN/dS below 1 indicates negative selection pressure (relaxed purifying selection), while a value above 1 indicates positive selection (selection for diversity), and dN/dS equal to 1 indicates neutral selection38. The dN/dS ratios of all groups in the survey appeared to be close to 0, which meant that NPY-family receptors have undergone pronounced negative selection in the vertebrate lineage, implying that these genes have essential functions on the growth and survival of organisms39.
Structure analysis of the NPY receptors in allotetraploid common carp
NPY-family receptors belong to the G-protein coupled receptor and have seven transmembrane domains (7tm_1), which facilitate the transmembrane transport of NPY and related peptides, thereby acting on the body’s peripheral tissues, and regulating growth, food intake, immunity and other physiological functions40. According to the result of secondary structure prediction, we found that all npyr genes in allotetraploid common carp have seven transmembrane domains (Supplementary Fig. 1). In addition, low complexity sequences were found at the N-terminal of two copies of npy4r, all of which confirmed the accuracy of common carp npyr gene identification.
The NPY-family receptors can be divided into three subfamilies based on amino acid sequence similarity and evolutionary origins, such as those of the Y1, Y2, and Y5 subgroups. The phylogeny trees of all npyr genes in allotetraploid common carp were constructed using their peptide sequences, showing the paralogous relationship of npyr gene copies and a clear cluster of two subtypes (Fig. 3a). Generally, the structure of the receptor protein is closely related to the affinity between receptor proteins and their related ligands. Motif analysis showed that most motifs of npyr genes in allotetraploid common carp are highly conservative (Fig. 3b).
Phylogenetic tree and motif analysis of NPY-family receptor genes in allotetraploid C.carpio. (a) The phylogenetic tree of NPY-family receptor genes in allotetraploid C. carpio. (b) Sequences analysis of NPY-family receptors in C.carpio, with colored pieces representing conservative motif patterns on these receptors. Secondary structural analysis of these sequences is shown in supplementary Fig. 1.
Nevertheless, we found that receptors of different subtypes have their unique motifs (Fig. 4). For Y1 subtype receptors, including npy1r, npy4r, npy8ar, npy8br, and their copies, there is a C-terminal specific motif 8. The logo distribution of motif 8 showed that the amino acid variants usually occur among different receptors rather than different copies of receptors (Fig. 4a). Furthermore, motif 9 is specific at the N-terminal of Y2 subtype receptors, while motif 10 is specific at the C-terminal, showing the structural differences between Y2 receptors (Fig. 4b,c).
Structure and motif characteristics of two subtypes of NPY-family receptors in allotetraploid C. carpio. (a) Y1 subtype receptors in allotetraploid C. carpio and their common unique motif. Motif 8 is unique to the Y1 receptor subtype of C. carpio. On the left are a logo representation and distribution of motif 8 for the C. carpio NPY-family receptors. On the right are examples of four Y1 receptors (Cca_npy1r-1, Cca_npy4r-1, Cca_npy8ar-2 and Cca_npy8br-2), showing the distribution of motif 8 in transmembrane domains. (b, c) Y2 subtype receptors in allotetraploid C. carpio and their common unique motifs. Motif 9 is specific to the N-terminal of Y2 subtype receptors, while motif 10 is unique to the C-terminal of Y2 subtype receptors. On the left are logos representing the distribution of motif 9 and motif 10 for the C. carpio NPY-family receptors. On the right are the transmembrane domains of an example of two Y2 receptors (Cca_npy2r-1, Cca_npy7r-2).
Chromosomal location and gene duplication of npyr genes
The allotetraploid C. carpio has two sets of subgenome A and B23. Comparing the C. carpio and its subgenome B progenitor-like diploid P. huangchuchieni with model species D. rerio, there were two copies of C. carpio npyr genes located on a pair of homologous chromosomes, which were derived from the two sets of subgenome A and B of the common carp genome, respectively (Fig. 5a). This implied that after the carp-specific fourth round of whole-genome duplication event (Cs4R WGD), npyr genes underwent a complete doubling and re-allocation of gene function in two subgenomes.
Chromosomal distribution and comparative expression profiles of NPY-family receptor genes from diploid reference species and subgenomes of allotetraploid C. carpio. (a) Chromosomal distribution and synteny plot of npyrs from three species of Cyprinidae. Gray lines indicate synteny blocks within diploid D. rerio and allotetraploid C. carpio and its B-subgenome progenitor-like diploid P. huangchuchieni, and red lines highlight the npyr gene pair. (b, c) The comparative expression profile between allotetraploid C. carpio subgenomes and its B-subgenome progenitor-like diploid P. huangchuchieni. The left heatmap shows subgenome comparative expression patterns of npyr genes during common carp embryo developmental stages, and the right heatmap displays the tissue-specific comparative expression profile of npyr genes among subgenomes of allotetraploid C. carpio and its B-subgenome progenitor-like diploid P. huangchuchieni, both with stage and tissue names listed on the right side of the panel, and all npyr gene names in C. carpio are listed at the bottom of the panel.
In addition, most npyr genes are evenly distributed among the ten paralogous chromosomes in C. carpio subgenomes, of which there are up to two npyr genes on chromosome A01 and B01. The six npyr genes of C. carpio subgenome B progenitor-like diploid P. huangchuchieni were located on five chromosomes, the same as in model species D. rerio, with npy1r and npy2r distributed on chromosome 1 together (Fig. 5a). The results showed that npy1r and npy2r in the three species of Cyprinidae are tandem duplication genes.
Comparative expression patterns of npyr genes
In order to reveal the comprehensive expression patterns of npyr genes in allotetraploid common carp, we analyzed and compared the gene expression levels at different embryonic developmental stages in its subgenomes (Fig. 5b). The results showed that most genes were preferentially expressed in sperm and egg, except that two copies of npy4r were specifically expressed in the egg stage, and there was no such obvious biased subgenome expression of npyr genes at early developmental stages41. In addition, the specific expression genes in each developmental stage that appeared after fertilization, such as npy8ar genes, showed specific expression in fertilized eggs, and it was speculated that the Y8a receptor might be able to regulate key downstream pathways in the early stages of embryonic development. Npy1r-2 and npy7r-1 were specifically expressed during the gastrula and muscle contraction stages, respectively. In summary, the results showed that the global expression of npyr genes in sperm and egg gradually became the dominant expression of specific genes at specific stages of embryonic development in allotetraploid common carp, which is the dominant gene in the common carp reproductive system. This might hint that the role of NPY-family receptors has been gradually redeployed in regulating growth and immune function in individual developmental processes.
RT-PCR results for different tissues in T. rubripes showed that npyr genes are expressed in the brain and also expressed in other peripheral tissues42. In order to investigate the tissue-specific expression patterns of npyr genes in allotetraploid C. carpio, we analyzed transcriptome data from twelve tissues of C. carpio and P. huangchuchieni. It was found that most of the npyr genes were expressed in the brain, skin, and gonad (Fig. 5c). In the brain, npy2r and npy4r were highly expressed in C. carpio subgenome A, but low or no expression in C. carpio subgenome B and its progenitor-like diploid P. huangchuchieni, while npy7r was not expressed in the brains of all three. For other tissues, we found that npy1r of C. carpio subgenome A was exclusively expressed in the spleen, which demonstrated a related form of immunity to the immune system in the common carp. Most npyr genes apart from npy8br were commonly expressed in the skin of C. carpio, but a similar situation did not occur in P. huangchuchieni, which implied a homogeneity within C. carpio genomes. In addition, npy8br in C. carpio subgenome B was specifically highly expressed in the gonad, while npy2r, npy4r, and npy7r genes of P. huangchuchieni were highly expressed, which indicated that the tissue expression pattern of C. carpio subgenome B was more similar to that of P. huangchuchieni. In general, the above results all supported the role of npyr genes in the growth and immunity of organisms43. Similar gene expression patterns also provided evidence that P. huangchuchieni may be a possible progenitor of C. carpio subgenome B.
Discussion
In this study, we investigated the NPY-family receptor genes of allotetraploid common carp and explored their evolutionary footprint and tissue distribution in different organisms. S. salar, O. mykiss and C. auratus who have experienced linage specific whole-genome duplications are thought to have led to the production of additional receptors and peptides in the NPY system44. We involved all the above species in this study, as well as the progenitor-like diploid species of allotetraploid C. carpio, such as P. huangchuchieni and O. macrolepis. The results showed that the number of npyr genes in tetraploid species was twice as much as that in diploid species. However, the distribution of the npyr genes in the autotetraploid and the diploid in the same habitat is not completely 2:1, which means that the gene loss may have occurred in S. salar and O. mykiss after the ancient whole-genome duplication event, but the gene copy produced by the recent polyploidy event in C. carpio and C. auratus has not changed much. This study elucidated the evolutionary fate of npyr genes after species-specific whole-genome duplication events in teleosts.
As is known, Y1 and Y5 receptors are usually considered as receptors for promoting food intake10. Taking Japanese medaka in marine habitat and zebrafish in freshwater habitat as model organisms, we found that Y1 receptor lost in teleost from marine habitat, but was detected in freshwater habitat species, and additional copies of Y1 receptor were found in goldfish, which may have experienced unique amplification14. In addition, a study on the expression profile of npyr genes in different tissues of common carp showed that Y1-1 was specifically expressed in the spleen45, suggesting the role of Y1 receptors in immunity5,43.
We found that the Y1 receptor was deficient in several fish in marine habitats but was present in all freshwater habitats. Whether the coincidence of distribution characteristics has anything to do with habitat remains unknown and needs further research to prove it. To explore the relationship between npy1r and the living environment of fish, we investigated the orthologues of npy1r in Ensembl, and a total of eleven fish had orthologues of this gene, of which ten were freshwater fish. Only Atlantic herring (Clupea harengus) was an exception. It lived in the Atlantic Ocean and was a type of marine migratory fish. The salinity range for a live environment is 5–35 ‰46. By constructing the species phylogenetic tree, we found that the genetic distance between this group and freshwater fish was closer, which might explain why, although it lived in seawater, it still had the npy1r gene, while other marine fish lacked this gene.
In addition, there were three copies of the Y1 receptor in goldfish, one more copy than in allotetraploid common carp, while there were only one copy of Y4 and Y7 in goldfish14,47. Since Y1 was the main feeding-promoting receptor, we believed that it might have additional functional differentiation and should be enhanced in goldfish. The npyr genes in common carp evenly distributed in two sets of subgenomes homologous to diploid species which confirmed the relatively new results of carp-specific polyploidy events23. The lack of copies of npyr genes in two ancient autotetraploid populations might be related to the early rediploidization19.
Because the Y5 receptor was not present in the teleost in this study, we suspected that this gene may have been lost in the evolution of teleost but retained in mammals and humans. Therefore, we investigated the orthologues of Y5 and found that they only existed in three ancient groups, Scleropages formosus, Erpetoichthys calabaricus and Lepisosteus oculatus which supported our conclusions. Up to now, Y1 and Y5 receptors have been ancient NPY receptor relics, which was confirmed in study1,11. Y5 existed in the early basal teleost but was lost in modern teleosts, whose genome became more complicated over time. In contrast, however, the third and fourth whole-genome duplication events from fish did not show any trace of the NPY5R gene.
Considering the allotetraploid background of C. carpio, the additional fourth round of whole-genome duplication events added to its genomic complexity. The gene copies generated through whole-genome duplication usually have various evolutionary fates, such as neofunctionalization, subfunctionalization, pseudogenization and gene loss, and functional conservation48. The evolutionary trend of the endocrine system of allotetraploid C. carpio after the fourth round of whole-genome duplication is an interesting topic, and existing research from our laboratory has found that the expression pattern of taste receptor genes indicated extensive gene functional differentiation49. The olfactory receptor gene family evolved asymmetrically between subgenomes, and the gene loss may have resulted in the reduction of the number and diversity of olfactory receptor genes50. Further in this study, our results showed that the npyr genes of allotetraploid common carp had an expressed preference among two subgenomes, and the B subgenome had a similar expression pattern with its diploid progenitor-like P. huangchuchieni. These studies provide insights into the evolutionary dynamics of the endocrine system of C. carpio after the fourth round of whole-genome duplication events.
Conclusions
In conclusion, a comprehensive genomic identification of npyr genes in allotetraploid common carp and nine other representative species was conducted to display the landscape of npyr genes in vertebrates. Detailed information on the multiple alignments, phylogeny relationship, conservative motifs and structural analysis revealed two subtypes of twelve npyr genes in allotetraploid common carp. Gene tandem duplication and chromosomal location of npyr genes were performed via syntenic analysis to decipher the evolutionary process of allotetraploid common carp after the fourth round of whole-genome duplication events. Furthermore, the comparative expression profiles of embryonic development in allotetraploid common carp showed an expression preference between two subgenomes. In adult tissues, the expression pattern of common carp was similar to C. carpio subgenome B progenitor-like diploid P. huangchuchieni. Overall, a full genomic investigation and comparative expression profiles of npyr genes in allotetraploid C. carpio and related species could provide new insight into future exploration of the function of npyr genes and the neuroendocrine system of allotetraploid C. carpio and vertebrates.
References
Larhammar, D. & Bergqvist, C. Ancient grandeur of the vertebrate neuropeptide Y system shown by the Coelacanth Latimeria chalumnae. Front. Neurosci. https://doi.org/10.3389/fnins.2013.00027 (2013).
Wan, Y. et al. Molecular characterization of CART, AgRP, and MC4R genes and their expression with fasting and re-feeding in common carp (Cyprinus carpio). Mol. Biol. Rep. 39, 2215–2223. https://doi.org/10.1007/s11033-011-0970-4 (2012).
Shiozaki, K. et al. Neuropeptide Y deficiency induces anxiety-like behaviours in zebrafish (Danio rerio). Sci. Rep. 10, 5913. https://doi.org/10.1038/s41598-020-62699-0 (2020).
Reichmann, F. & Holzer, P. Neuropeptide Y: A stressful review. Neuropeptides 55, 99–109. https://doi.org/10.1016/j.npep.2015.09.008 (2016).
Gonzalez-Stegmaier, R. et al. New immunomodulatory role of neuropeptide Y (NPY) in Salmo salar leucocytes. Dev. Comp. Immunol. 76, 303–309. https://doi.org/10.1016/j.dci.2017.06.018 (2017).
Assan, D. et al. The roles of neuropeptide Y (Npy) and peptide YY (Pyy) in teleost food intake: A mini review. Life 11, 547 (2021).
Salaneck, E., Larson, E. T., Larsson, T. A. & Larhammar, D. Effects of a teleost tetraploidization on neuropeptide Y receptor gene repertoire in ray-finned fishes. Ann. N. Y. Acad. Sci. 1040, 457–459 (2005).
Larhammar, D. & Salaneck, E. Molecular evolution of NPY receptor subtypes. Neuropeptides 38, 141–151. https://doi.org/10.1016/j.npep.2004.06.002 (2004).
Salaneck, E., Larsson, T., Larson, E. T. & Larhammar, D. Birth and death of neuropeptide Y receptor genes in relation to the teleost fish tetraploidization. Gene 409, 61–71 (2008).
Larsson, T. A. et al. Pufferfish and zebrafish have five distinct NPY receptor subtypes, but have lost appetite receptors Y1 and Y5. Ann. N. Y. Acad. Sci. 1040, 375–377 (2005).
Larsson, T. A., Larson, E. T. & Larhammar, D. Cloning and sequence analysis of the neuropeptide Y receptors Y5 and Y6 in the coelacanth Latimeria chalumnae. Gen. Comp. Endocrinol. 150, 337–342 (2007).
Larsson, T. A., Larson, E. T., Fredriksson, R., Conlon, J. M. & Larhammar, D. Characterization of NPY receptor subtypes Y2 and Y7 in rainbow trout Oncorhynchus mykiss. Peptides 27, 1320–1327 (2006).
Sundstrom, G., Larsson, T. A., Brenner, S., Venkatesh, B. & Larhammar, D. Evolution of the neuropeptide Y family: New genes by chromosome duplications in early vertebrates and in teleost fishes. Gen. Comp. Endocrinol. 155, 705–716. https://doi.org/10.1016/j.ygcen.2007.08.016 (2008).
Narnaware, Y. & Peter, R. Neuropeptide Y stimulates food consumption through multiple receptors in goldfish. Physiol. Behav. 74, 185–190 (2001).
Batterham, R. L. et al. Gut hormone PYY 3–36 physiologically inhibits food intake. Nature 418, 650–654 (2002).
Fredriksson, R., Larson, E., Yan, Y., Postlethwait, J. & Larhammar, D. Novel neuropeptide Y Y2-like receptor subtype in zebrafish and frogs supports early vertebrate chromosome duplications. J. Mol. Evol. 58, 106–114 (2004).
Larsson, T. A. et al. Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol. Biol. 8, 184. https://doi.org/10.1186/1471-2148-8-184 (2008).
Allendorf, F. W. & Thorgaard, G. H. Evolutionary Genetics of Fishes 1–53 (Springer, 1984).
Lien, S. et al. The Atlantic salmon genome provides insights into rediploidization. Nature 533, 200–205 (2016).
Xu, P. et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 46, 1212–1219 (2014).
Li, Y. et al. Construction and characterization of the BAC library for common carp Cyprinus carpio l and establishment of microsynteny with zebrafish Danio Rerio. Mar. Biotechnol. Mar. Biotechnol. 13, 1183–1183 (2011).
Xu, P., Wang, J., Wang, J., Cui, R. & Sun, X. Generation of the first BAC-based physical map of the common carp genome. BMC Genom. 12, 537 (2011).
Xu, P., Xu, J., Liu, G., Chen, L. & Sun, X. The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat. Commun. 10, 1–11 (2019).
Chen, L. et al. Chromosome-level genome of Poropuntius huangchuchieni provides a diploid progenitor-like reference genome for the allotetraploid Cyprinus carpio. Mol. Ecol. Resour. 21, 1658–1669 (2021).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. https://doi.org/10.1093/bioinformatics/btm404 (2007).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. https://doi.org/10.1093/nar/gku316 (2014).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. https://doi.org/10.1093/molbev/msy096 (2018).
Edgar, R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. https://doi.org/10.1093/nar/gkh340 (2004).
Subramanian, B., Gao, S., Lercher, M. J., Hu, S. & Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. https://doi.org/10.1093/nar/gkz357 (2019).
Elkan, T. L. B. A. C. in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology.
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Omasits, U., Ahrens, C. H., Müller, S. & Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. https://doi.org/10.1093/bioinformatics/btt607 (2013).
Kryazhimskiy, S. & Plotkin, J. B. The population genetics of dN/dS. PLoS Genet. 4, e1000304. https://doi.org/10.1371/journal.pgen.1000304 (2008).
Delport, W., Poon, A. F., Frost, S. D. & Kosakovsky Pond, S. L. Datamonkey 2010: A suite a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457. https://doi.org/10.1093/bioinformatics/btq429 (2010).
Wang, Y. et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49 (2012).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Nei, M. & Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986).
Persi, E., Wolf, Y. I. & Koonin, E. V. Positive and strongly relaxed purifying selection drive the evolution of repeats in proteins. Nat. Commun. 7, 13570. https://doi.org/10.1038/ncomms13570 (2016).
Gao, S. et al. Molecular characterization of neuropeptide Y (NPY) receptors (Y1, Y4 and Y6) and investigation of the tissue expression of their ligands (NPY, PYY and PP) in chickens. Gen. Comp. Endocrinol. 240, 46–60. https://doi.org/10.1016/j.ygcen.2016.09.005 (2017).
Luo, J. et al. From asymmetrical to balanced genomic diversification during rediploidization: Subgenomic evolution in allotetraploid fish. Sci. Adv. 6, 1. https://doi.org/10.1126/sciadv.aaz7677 (2020).
Campbell, R. K., Satoh, N. & Degnan, B. M. Piecing together evolution of the vertebrate endocrine system. Trends Genet. 20, 359–366. https://doi.org/10.1016/j.tig.2004.06.005 (2004).
Chen, W. C. et al. Neuropeptide Y is an immunomodulatory factor: Direct and indirect. Front. Immunol. https://doi.org/10.3389/fimmu.2020.580378 (2020).
Sundström, G., Larsson, T., Xu, B., Heldin, J. & Larhammar, D. Interactions of zebrafish peptide YYb with the neuropeptide Y-family receptors Y4, Y7, Y8a, and Y8b. Front. Neurosci. https://doi.org/10.3389/fnins.2013.00029 (2013).
Jiang, Y. et al. Transcriptome signatures in common carp spleen in response to Aeromonas hydrophila infection. Fish Shellf. Immunol. 57, 41–48 (2016).
Limborg, M. T. et al. Environmental selection on transcriptome-derived SNPs in a high gene flow marine fish, the Atlantic herring (Clupea harengus). Mol. Ecol. 21, 3686–3703. https://doi.org/10.1111/j.1365-294X.2012.05639.x (2012).
de Pedro, N. et al. NPY receptors and opioidergic system are involved in NPY-induced feeding in goldfish. Peptides 21, 1495–1502 (2000).
Glasauer, S. M. K. & Neuhauss, S. C. F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 289, 1045–1060. https://doi.org/10.1007/s00438-014-0889-2 (2014).
Kong, S. et al. Genome wide identification of taste receptor genes in common carp (Cyprinus carpio) and phylogenetic analysis in teleost. Gene 678, 65–72 (2018).
Wang, H. et al. Genome-wide identification and characterization of olfactory receptor genes in common carp (Cyprinus carpio). Gene 777, 145468 (2021).
Funding
This work was supported by the National Key R&D Program of China (2019YFE0119000), and the National Natural Science Foundation of China (31872561).
Author information
Authors and Affiliations
Contributions
P.X. conceived the project and P.X. contributed to the funding acquisition. X.Q.Z. wrote the manuscript and performed all analyses, while L.C. produced the RNA-seq data. B.J.L. and J.Z.X. helped with the revision of this paper. All authors have validated the final manuscript and appreciated the quality of it as it was published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zou, X., Chen, L., Li, B. et al. The neuropeptide Y receptor gene repository, phylogeny and comparative expression in allotetraploid common carp. Sci Rep 12, 9449 (2022). https://doi.org/10.1038/s41598-022-13587-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-13587-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.