Abstract
Cytokinesis block micronucleus (CBMN) assay is a widely used radiation biological dose estimation method. However, the subjectivity and the time-consuming nature of manual detection limits CBMN for rapid standard assay. The CBMN analysis is combined with a convolutional neural network to create a software for rapid standard automated detection of micronuclei in Giemsa stained binucleated lymphocytes images in this study. Cell acquisition, adhesive cell mass segmentation, cell type identification, and micronucleus counting are the four steps of the software's analysis workflow. Even when the cytoplasm is hazy, several micronuclei are joined to each other, or micronuclei are attached to the nucleus, this algorithm can swiftly and efficiently detect binucleated cells and micronuclei in a verification of 2000 images. In a test of 20 slides, the software reached a detection rate of 99.4% of manual detection in terms of binucleated cells, with a false positive rate of 14.7%. In terms of micronuclei detection, the software reached a detection rate of 115.1% of manual detection, with a 26.2% false positive rate. Each image analysis takes roughly 0.3 s, which is an order of magnitude faster than manual detection.
Similar content being viewed by others
Introduction
Once large-scale radiation accident occurs involving lot of people, it is critical to evaluate their exposure dose for further treatment1, so as to determine the priority for treatment and alleviate their unnecessary worries. Since ionizing radiation can cause chromosome damage, the biological dose can be estimated by cytogenetic methods. Dicentric chromosome detection2 is the most reliable cytogenetic dose estimating approach, but the analysis process is very time-consuming and requires excellent professional skills and knowledge. Therefore, when a large number of samples need to be evaluated, other dose estimating methods3,4,5,6,7 have aroused the interest of researchers. Among these other methods, CBMN analysis8,9 is the best alternative method.
CBMN analysis is a way of producing binucleate cells by adding a proper quantity of cytochalasin B to the cell culture process, causing the nucleus to divide normally while the cytoplasm does not. Since micronucleus (MN) is a fragment or an entire the chromosome, the binucleated cell's MN rate can be correlated with the exposure dose to estimate the unknown dose10,11,12,13, similar to the detection of chromosome aberrations. When the cell is irradiated, MN is in a hysteresis condition in the late stages of nuclear division. As a result, MN does not reside in the main nucleus of the cell, but rather forms a tiny circular entity and remains in the cytoplasm14. Compared with dicentric chromosome detection, CBMN analysis is easier and faster, making it is more appropriate for rapid biological dosimetry in a large number of persons. In addition to radiation dose estimation, CBMN analysis is also an effective detection method for studying the potential genotoxicity and cytotoxicity of chemicals15,16. Due to the subjectivity and long assay time of manual detection17,18, manual detection cannot satisfy the demands of rapid standard detection. To overcome these problems and produce superior statistical data and reproducibility, automated detection of MN has become a critical topic.
Currently, there are a variety of automated analysis methods for detecting binucleated cells (BNCs) and micronuclei (MNi). One of the most important distinctions between automated analysis algorithms is whether or not the studied object comprises cytoplasm. The first sort of algorithm only analyzes just the nucleus information and does not correlate the cytoplasmic information of related cells19,20,21,22,23,24,25,26,27,28,29, which is suitable for images captured by automated microscopes19,20,21,22,23 and imaging flow cytometers24,25,26,27,28,29. When employing imaging flow cytometers, only fluorescent dyes can be used to mark cell nuclei and micronucleus, and the detection method can be the conventional gating strategy27 or the deep learning CNN algorithm28,29. When using automatic microscopes, both fluorescent dye and Giemsa dye can be utilized for cell staining. Since Giemsa marks both the nucleus/MN and the cytoplasm, it will have an inescapable effect on the nucleus/MN recognition. Therefore, fluorescent staining is frequently favored when employing automatic microscopes20,21. This algorithm uses a threshold algorithm to determine the location of the cell nucleus, then the distance between each nucleus to determine the location and range of the cell, and finally the conventional gating strategy or a pre-trained convolutional neural network (CNN) model to determine the type of cell and the number of MNi.
The second kind of algorithm makes the entire cell as the detecting unit30,31,32,33,34,35,36,37. This algorithm is more in line with BNC detection criteria38 and is appropriate for images captured by automatic microscopes. When it comes to cell staining, the dye must not just stain the nucleus/MN, but the entire cell as well. The visualized nucleus and cytoplasm can be used to match the nucleus/MN with the corresponding cells to identify BNCs and MNi more accurately. This algorithm separates the image backdrop, cytoplasm, and nucleus/MN using different pixel thresholds, then employs a gating strategy to determine the cell's integrity and the number of nuclei/MNi30,31. Because the staining residue of Giemsa can cause blurred cytoplasmic background and produce some artifacts similar to MN, some researchers stain the nucleus and cytoplasm separately with two fluorescent dyes32 or use higher magnification imaging technologies to reduce the false positive incidence of MN35,36.
The current study has demonstrated that automatic detection of MN has engineering application value, but existing algorithms still have some drawbacks, such as low MN detection rates. In this study, a rapid standard automatic analysis software for Giemsa stained binuclear lymphocyte images based on automatic microscope imaging was designed using CNN. Low hardware costs and repeatable slide scanning are two advantages of automated microscopes. Giemsa dyes are economical, and the slides can be kept for years. Furthermore, the algorithm can distinguish BNCs and MNi even when the cytoplasm is hazy, and it can also detect monocytes (MOCs) and multinucleated cells (POCs).
Materials and methods
Software and hardware platform
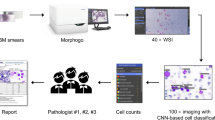
To implement rapid CBMN analysis, high-definition microscopic images of the cells must be obtained at the very beginning. The image acquisition system is made up of three parts (Fig. 1): (1) A Color microscope (OLYMPUS, Japan) with 10x, 20x, and 40× objective lenses. (2) A CCD camera (Lumenera, Canada), which is connected to the computer through the USB interface to allow for real-time display and picture taking. (3) A computer and a motion platform.
The computer sends commands to the motion console through Ethernet to move it along the predefined path. The microscope travels up and down to acquire photos that are then saved in the computer. 3200 pictures can be captured by scanning an entire standard-size glass slide using a 10× eyepiece and a 20× objective lens, depending on the camera magnification and the size of the normal glass slide.
On a desktop computer with a 6-core Intel Core i7-8700K 3.70 GHz CPU (32G RAM) and NVIDIA GeForce GTX 1080 Ti GPU (11G memory), software development and picture analysis were carried out. The software development tool uses Visual Studio 2013 and PyCharm 2018. Image capture, image processing, image analysis, and data storage management are among the four modules included in the software. Open CV and other third-party libraries are included in the software support library. C++ uses Python scripts to realize the categorization of distinct sorts of cells.
Slide source
China Henan Institute of Occupational Disease Prevention and Control was entrusted for manufacturing 100 samples of CBMN. All donors are nonsmokers who have been maintained generally well at the time of blood collection, have no visible ailments such as the common cold, flu, or pneumonia, and have not been exposed to medical treatment in the previous 12 months. Informed consent was obtained from all subject and/or their legal guardian. A 4 Gy gamma irradiation was applied to all blood samples.
All experimental protocols of this research submitted in Scientific Reports were approved by the Ethic Committee of the China Henan Institute of Occupational Disease Prevention and Control and conformed to the Declaration of Helsinki, including:
-
Collection of human blood.
-
Cell culture and glass slide production.
-
Collection of microscopic cell images.
-
Automated detection and manual analysis of binuclear lymphocyte images.
The slide samples were prepared in compliance with the standards and criteria set out by the International Atomic Energy Agency2.
Image acquisition
The sample was imaged using an OLYMPUS microscope with a 20 × objective. Each field of vision received one image, which had a size of 2048 × 2048 pixels and was saved in BMP format. There were 100 slides marked for model training and testing in total. 80 out of 100 slides from 16 donors (8 males, 8 females) were used for CNN model training, whereas the residual 20 slides from the other 4 donors (2 males and 2 females) were kept untouched until the final testing. The training-marked slides were scanned and photographed by our system.
Design
There are four phases in the picture analysis process. First, extract the cells from the image background. Then, segment individual cells from the adhesive cell masses. Then, use the trained CNN model to detect and categorize individual cells. Finally, count the number of MNi of the BNC.
Cell acquisition
Threshold segmentation is done on the acquired picture initially to generate a full cell image. The original color image (Fig. 2A) was transformed into grayscale images of R, G, and B channels according to the RGB channels before threshold segmentation. The image of the sample will seem dark blue or reddish purple in human vision since it has been stained with Giemsa. As a result, the cytoplasm/nucleus pixel intensity in the G channel grayscale picture (Fig. 2B) is the highest among the four images of R, G, B grayscale images and conventional grayscale photos. The picture is then thresholded using the iterative thresholding approach for the G-channel gray image. Each image (2048 × 2048 pixels) takes roughly 0.2 s to process using the iterative threshold classification algorithm. The mask image is eroded and extended after thresholding to eliminate certain minor noise spots and contaminants. Finally, the mask image is combined with the original color image, as seen in Fig. 2C.
Classification of individual cells and adhesive cell masses
Since the cell distribution on the glass slide is not uniform, sticky cell masses will appear in the photos produced in III.A. To improve BNC detection rates, the adhesive cell mass must be segmented into individual cells. Before segmentation, all of the acquired cells must be classified into individual cells and adherent cell clusters.
There are two aspects of the classification method that are evaluated: 1. Each cell should have a complete cytoplasm and be nearly round; 2. The area value of the cell should be within a reasonable range. Based on these issues, cell area, cell extension rate, and cell defect rate are used as judgment variables.
(1) The cell area threshold is used to filter out tiny impurities and big adherent cell clusters. These impurities or cell clusters will significantly increase the algorithm's analysis time.
The area threshold's lower limit is about the minimum area value of a cell (1000 pixels, which is comparable to around 100 μm2), while the upper limit is approximately the area value of 5 cells (8000 pixels) (when the number of adhering cell clusters is more than 5, the adhesion situation is complicated and the cell detection rate of the watershed algorithm is low.). These variables can remain constant after the microscope magnification and camera specifications have been determined. A series of thresholds have been tested as shown in Supplementary Table S2.
(2) The calculation formula of the elongation rate parameter is:
where H is the width of the cell's smallest circumscribed rectangle and W is the length. The elongation rate is a measurement of how near an object's form is to a circle. The elongation value of a circle is 1; for things that are not as round, the elongation value is less than 1, and as the form of the item approaches non-circular, the elongation value approaches 0.
(3) The formula for calculating the defect rate is:
where A is the area of the cell's smallest circumscribed circle, and B is the cell's total area. The elongation rate indicates how close the object's real area is to the circular area. The defect rate for a circle is 0; for cell objects with incomplete cytoplasm, the defect rate is higher.
The elongation rate and defect rate thresholds are used to separate the images into two types: individual cells and adherent cell clusters (the impurities will be further identified in the CNN model). A large number of images will be assessed as adhesion cell clusters if the parameter setting is too stringent (for example, the elongation rate is 0.8–1.0). The watershed technique will be used to segregate these images, slowing down the algorithm's analysis pace and lowering the BNC detection rate (some intact cells are over-segmented). If the parameters are set too loosely (for example, 0.2–1.0), many adherent cell clusters that need to be separated will be evaluated as individual cells, reducing the number of cells found by the algorithm. A variety of thresholds were tested in the process of defining parameter thresholds, as shown in Supplementary Table S1.
The two judgment parameters are combined in a "parallel" way, which means that when an item's elongation rate parameter and defect rate parameter are both within the allowed range, the object is regarded as an individual cell. When the values in the group 4 of Table S2 (the Elongation rate is set to 0.5–1.0, the Defect rate is set to 0–0.5, and the area threshold is set to 1000–8000) are used, it is more consistent with the definition of intact cells38, and an ideal balance between the algorithm's cell detection rate and the analysis speed can be achieved.
Adhesive cell masses segmentation
The adhesion cell masses were segmented using the watershed algorithm39 in this step. To get the distance gradient map, first grayscale the image of the sticky cell mass, and then continually corrode the grayscale image. Finally, the edge lines of each cell in the image of the sticky cell mass are obtained using the watershed method on the distance grayscale image.
The use of traditional watershed algorithm will result in excessive segmentation since the form and size of the cells varies, and certain contaminants and cells stick together. As a result, the seed point area threshold judgment was added to the algorithm. To avoid excessive segmentation, the number of pixel points of the seed point was used as the criteria, and spurious seed points with too tiny area values were rejected. The lowest seed point threshold was set at 100 pixels in this algorithm. Figure 3 shows the adhesion cell cluster segmentation findings.
Cell type recognition based on CNN model
CNN is made up of image input layer, convolutional layer, maximum pool, average pool, comprehensive connection layer, and other components as a deep learning algorithm. It employs mapping to take the original pixels of the input picture as input, extracts features in a hierarchical way, and then classifies the features using a fully connected layer. The number of layers can be raised or lowered depending on the size of the input picture, and it is not required to use all of the layers in the network. CNN has the benefit of not requiring any artificial feature extraction using other approaches.
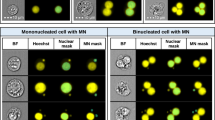
A CNN model for image categorization of micronucleus cells is developed, and the suggested CNN architecture is shown in Fig. 4. There are seven layers in this CNN model (4 convolutional layers, 3 fully connected layers). The first convolutional layer (Conv1) uses 8 kernels with a size of 3 × 3 to filter a 3 × 132 × 132 input image. Conv2 has 16 filters with a size of 3 × 3, and Conv 3 and Conv 4 have 32 and 48 filters with a size of 3 × 3, respectively. The first fully connected layer (FC1) has 40 neurons, and each neuron uses a 50% loss rate during training. FC2 has 20 neurons, and each neuron also uses a 50% loss rate during training. After Conv1, Conv2, and Conv4, the largest pooling layer with a 2 × 2 kernel size is used. The rectified linear unit (ReLU) activation function is used after each convolution and fully connected layer. In the end, the softmax function is used to generate probability distributions on the five class labels.
On the images of 80 slides used for training, about 280,000 images of individual cells were produced after conducting cell extraction and segmentation of adhesion cell clusters. Five experienced analysts carried out the categorization according to defined guidelines38. Cell images were divided into five categories: 0. BNC without MN, 1. Binucleated cell with Micronuclei (BNCMN), 2. MOC, 3. POC with three nuclei and above, 4. Any other cell or substance that does not belong to the first four types. 24,000 BNC images without MN, 16,000 BNCMN images, 45,000 MOC images, 35,000 POC images, and 160,000 other types of images were obtained in the end. The amount of BNC and BNCMN photos is only 14% of the total number of images, and the number of different types of images is not evenly distributed. To increase the model's capacity to detect BNC/BNCMN images, the BNC/BNCMN images are oversampled by 2 times and 4 times, respectively. In addition, to balance the corresponding category photos in the training set, data augmentation methods were utilized to produce new images. Horizontal flip, random addition of noise or blocks with occlusion effects on the original picture, main component assay (PCA) dimensionality reduction processes are some of the techniques employed. Two dropout layers were inserted between the FC1 and FC2 layers (dropout = 0.5) and between the FC2 and FC3 layers (dropout = 0.5) of the model to reduce over-fitting caused by picture over-sampling. According to a 7:2:1 ratio, each type of picture was randomly sorted into a CNN training set, validation set, and test set. The training and validation sets are involved in the CNN model training process, but the test set is not involved in the model training and just acts as a preliminary internal test.
Since the area values of individual cell pictures varies, all photos must be scaled to a 132 × 132 sized image without stretching before training (132 × 132 is close to the average area of BNC images). Then use TensorFlow's Keras function package to implement the CNN model. The learning rate was initialized to 0.001 and reduced by a factor of 10 after half of the iterations. The momentum term was set to 0.9, and the weight decays was set to 0.0001. All parameter choices, with the exception of the given configuration, are standard setups. The network was trained on the NVIDIA GeForce GTX 1080 Ti GPU for 60 iterations in 40 h. The model's convergence was good during the training phase, and there was no noticeable over-fitting problem. In the end, the model's total recognition accuracy was 85%, while the loss rate was around 0.4.
The trained model was internally tested using the images of the test set. BNC images without MN had a recognition accuracy of 84.5%, and BNCMN images had a recognition accuracy of 87.1%. Figure 5 depicts the test results for 5 different categories.
Micronucleus count
The CNN model in III.D can only identify cell types, but it didn't determine how many MNi are in a BNC with several MNi. Therefore, the number of MNi must be determined at this step of the work. The nucleus and MNi were extracted from the cytoplasm first, and then the numerous MNi that were joined attached and the MNi that were attached to the nucleus were segmented. Finally, the nuclei and MNi were sorted by area value to count the number of MNi.
The iterative threshold technique cannot properly extract nuclei and MNi from these images since there are many images of cells with hazy cytoplasm in the gathered images. As a result, a k-means clustering-based threshold segmentation technique was developed and Fig. 6 depicts the flowchart. Pure black (image background), light purple (cytoplasm), and dark purple (nucleus/ MN) are the three hues in the cell picture. The black background has a gray value of 0, the nucleus/MN has a gray value near to 0, and the cytoplasm has a gray value greater than 0. K was set to 2 to guarantee that the clustering method can successfully differentiate the cytoplasm from the nucleus/MN since the gray value difference between the black background and the nucleus/MN is lower than the difference between the nucleus/MN and the cytoplasm.
After extracting the nuclei and MNi, import them into the watershed algorithm. The corrosion parameters of the watershed algorithm used here are different from the watershed algorithm in III.C. Because the goal of III.C is to segment the adhesive cell masses, a higher corrosion parameter (value: 7) is required. The major purpose in this stage is to separate the adhesive MN masses into independent MNi, not to divide the adhering nucleus masses into independent nuclei (since the number of nuclei has already been determined). To intensify the segmentation effect, the watershed algorithm utilizes a smaller corrosion parameter (value: 3) in this stage. Figure 7 depicts the segmentation effect. The area value and coordinate information of each nucleus/MN can be acquired when the segmentation is complete.
When the area information of the cell nucleus and MNi have been obtained, they are ordered numerically.
Following that, the potential MNi are screened out based on the assumption that the maximum permitted MN area is 20% of the nuclear area value (if there are several nuclei, the average value of multiple nuclei is chosen). Finally, compute the number of MNi based on the premise that the smallest allowed MN area is 20 pixels (to prevent some noise points from being misjudged as MN).
Results
Random image algorithm test
Twenty slides from 4 separate donors (2 men and two females, A-D) were scanned and photographed, and the photos were then utilized to test the system. These 20 slides' images, as noted in II.C, did not take part in the CNN model's training. Following the completion of the image collection, 100 images were chosen at random from each slide's picture collection, 500 images for each donor, for a total of 2000 images.
The proofreaders also performed artificial visual scoring on 2000 original images according to the standard method38. The image analysis data obtained is shown in Table 1. Five experienced inspectors detected 1949 BNCs without MN, 1029 BNCsMN, and 1359 MNi, as indicated in Table 1. In all, the manual detection required around 7.5 h of proofreading time. These images are then automatically analyzed by the software, and the results are displayed in Table 2.
The automatic image analysis took roughly 620 s in total (about 0.3 s per image), which is 44 times faster than manual detection. 2041 BNCs without MN, 920 BNCsMN, and 1564 MNi were recognized by the software.
Following the automatic analysis of these images, the same proofreaders utilize the system's gallery to visually proofread the BNC images acquired from the automated analysis and count the result's error scores.
Table 3 shows the proofreaders' right diagnosis. Table 4 shows the relationship between automatically scored data and proper diagnosis by observers.
For model estimation, the quality parameters (Accuracy, Sensitivity, Specificity, Positive Predictive Value, Negative Predictive Value) were utilized. These parameters can be calculated via
TP: Ture Positive; FP: False Positive; FN: False Negative; TN: Ture Negative.
Table 5 shows the right diagnosis made by proofreaders. Table 6 shows the relationship between the gating strategy's automated scoring data and proper diagnosis by observers.
It has been found that utilizing an automated analysis approach based on the CNN model, the identification rate of BNC is significantly higher than using a conventional image recognition algorithm (99.4% vs 58.0%). Since most BNCs with two sticky cores are discarded, conventional image recognition algorithms achieve a high PPV (91.9% vs 85.3%) by systematically eliminating potential targets. As compared to the conventional gating strategy algorithm, the algorithm based on CNN tends to behave more adventurous (high TPR and low TNR).
There are no adjustments for age, sample storage duration, or the treated or untreated instances of patients, since the difference between cases and controls is not the major purpose of this study. And it has no effect on the direct comparison of visual and automated counting.
Assay of test results by SPSS
For studying the difference between the results of automatic analysis and manual detection, SPSS (Statistical Product and Service Solutions, version 19.0) was used to perform statistical analysis on the MN rate (MNi/BNCs) data in Tables 1 and 2. The 20 MN rates in Table 1 were regarded as the "Manual" group, and the 20 MN rates in Table 2 were regarded as the "Automatic" group. Since the amount of data is not large, and the two sets of data do not obey the normal distribution, the Mann–Whitney U was first used to test whether there is a statistical difference between the MN rate data of manual visual inspection and automatic analysis.
In the Mann–Whitney U test results, the average rank of the manual detection data is 13.30, and the average rank of the automatic analysis data is 27.70. The exact P value is < 0.001 (The specific data can be viewed in Supplementary Fig. S1). As a result, the statistical significant difference between manual detection and automatic analysis was determined. According to calculations, it can be found that the mean and median values of the data of the automatic analysis group are higher than the manual detection group, indicating that the value of the automatic grouping is generally higher than the manual detection group.
Then, the 20 MN rates in Table 1 were divided into four groups from A to D according to their respective donors (the number of samples in each group is 5). Since the amount of data is not large, and each group of data does not follow a normal distribution, the Kruskal–Wallis H test is performed. On the one hand, the Kruskal–Wallis H test can detect whether there are statistically significant differences between the 4 groups of data. On the other hand, two conclusions can be drawn using the Kruskal–Wallis H test on the MN rate data of manual detection and automatic analysis. The consistency between the two conclusions may then be shown. If both inspection conclusions are the same, it proves that the automatic analysis has the detection ability roughly equivalent to the manual detection. The test result data of the MN rate of manual detection is shown in Fig. 8.
It can be found that the distribution of the manually detected MN rate data is not the same, and the difference is statistically significant (P = 0.039 < 0.05, in Supplementary Fig. S2). The average rank of Donor A group data is 4.50, the average rank of Donor B group data is 10.20, the average rank of Donor C group data is 12.50, and the average rank of Donor D group data is 14.80.
After the significance level was adjusted by Bonferroni method, the data of each group were compared in pairs, and it was found that the data distribution of Donor A group and Donor D group (P = 0.035 after adjustment, P < 0.05) was statistically different, while the difference between the other groups was not statistically significant (the comparative data of other groups can be viewed in Supplementary Fig. S3). Similarly, the 20 MN rate data in Table 2 are also divided into four groups from A to D (n = 5) according to their respective donors. The test results are shown in Fig. 9.
It can be found that the distribution of the data of each group of the automatic analysis result is not completely the same, and the difference is statistically significant (P = 0.042 < 0.05, in Supplementary Fig. S4). The average rank of Donor A group data is 4. 20, the average rank of Donor B group data is 11.20, the average rank of Donor C group data is 12.40, and the average rank of Donor D group data is 14.20. After the significance level was adjusted by Bonferroni method, the data of each group were compared in pairs, and it was found that the data distribution of Donor A group and Donor D group (P = 0.045 after adjustment, P < 0.05) was statistically different, while the difference between the other groups was not statistically significant (The comparative data of other groups can be viewed in Supplementary Fig. S5).
In Fig. 8, the value next to the dot represents the average rank of the group. The connecting line represents the result of the pairwise comparison, the black connecting line indicates the difference between the two groups is not statistically significant, and the orange connecting line indicates the statistical significance of the difference between the two groups.
The conclusions of the Kruskal–Wallis H test for manually detected and automatically analyzed data are same. Both conclusions concluded that the data distributions of Donor A and Donor D are statistically significantly different. As a result, the automatic CBMN analysis algorithm has a detection capacity that is roughly equivalent to manual detection.
Discussion
The purpose of this research is to design a system that can automatically analyze Giemsa stained CBMN images and analyze the images while capturing. This method, unlike conventional gating strategy analysis algorithms, preserves cells to a higher extent, allowing it to detect more BNCs/MNi and be more adventurous. Conventional image recognition steps like manual feature extraction, dimensionality reduction, and feature ranking are also not needed. Because CNN can intelligently alter network settings to get the greatest classification performance and extract components autonomously by producing feature maps after each convolutional layer, allowing it to find the best performance elements on its own. A K-means clustering-based cell nucleus/MN extraction approach is developed to overcome the problem of CNN being unable to count MNi of BNCs with multiple MNi. The algorithm can also handle the adherence of numerous MNi to one another as well as the adhesion of MNi to the nucleus. The automatic CBMN analysis method detected BNCs and BNCsMN at 99.4% and 89.4% of manual detection, respectively, with BNCs false positive rates of 14.7%.
The most of pseudo-BNCs come from cells with overlapping complicated nuclei. When confronted with such images, even seasoned observers have difficulty determining whether the nuclei overlap or connect in space. MN had a detection rate of 115.1% as compared to manual detection, and a PPV of 73.8%. The most common source of false positives in MN is Giemsa dye, which causes a fuzzy cytoplasmic background and artifacts similar to MNi. These artifacts are also very difficult for manual detection. Some MNi with lighter hues, on the other hand, are overlooked by the algorithm since they are difficult to distinguish using the k-means clustering threshold approach. The identical Kruskal–Wallis H test result of manual detection and automatic analysis MN rate data demonstrates that the automatic CBMN analysis algorithm has detection capacity that is roughly equivalent to manual detection. The algorithm's detection accuracy and rate can be enhanced further by increasing the number of CNN training samples, eventually reaching a level that is totally comparable to that of experienced human observers.
Some algorithms19,20,21,22,23,24,25,26,27,28,29 in prior similar work only analyzed nuclear data. To establish the cell border, whether using the gating strategy19,20,21,22,23,24,25,26,27 or the CNN algorithm28,29, it is important to examine the relative position and size of the cell nucleus. When this type of algorithm is employed, it is typically required to prioritize the use of fluorescent stains20,21 in the slide production process and to limit cell density as much as possible to avoid the circumstance where some nuclei are accidentally assigned to the same cell22. The conventional gating strategy algorithm involves first searching the nucleus, then judging the two neighboring nuclei as a BNC, defining the circular region around the two nuclei of the BNC as the cell area, and then searching for MNi in this area. The gating strategy's determination parameters are based on morphological characteristics such as area22, circle26, overlap of two nuclei25, and so on, and this approach detects MN at a rate of 20–35%20,24,25 of manual counting. Other algorithms30,31,32,33,34,35,36,37 in prior similar work used the entire cell as the detection unit. This type of algorithm first identifies the cytoplasm of the cell to determine whether it is intact30,31,37, then detects the number of nuclei in the cell, and lastly counts the number of MNi. The MN detection rate analyzed by this sort of algorithm is about 65% of the manual count30,35. Our algorithm flow is similar to that of previous similar work that uses the entire cell as the detection unit. The detection rate of our system is higher than that of the prior system20,30 because it combines adhesion cell cluster segmentation and a binucleated cell identification strategy based on CNN, both of which were not included in previous similar work.
In terms of slide applicability and system clinical application, this system may obtain satisfactory identification results for slides created using conventional procedures without requiring high staining effect33 or cell density criteria31. The system can also detect BNCs and MNi with blurred cytoplasmic without utilizing a high microscope magnification, indicating that it has both great detection and speedy analysis capabilities. Unlike conventional image analysis algorithms, this software uses a CNN model for cell recognition. As a result, no tedious manual feature selection or threshold parameter adjustment is required, and all that is required is to manually give enough training data for CNN.
According to statistics, the system takes roughly 620s to analyze 2000 pictures, which is 44 times faster than manual detection. With enough BNCs in slide, the typical analysis time for biological dose estimation is just 7–10 min if completed according to the criteria of studying a minimum of 1000 BNCs. This unquestionably enhances the speed of CBMN analysis when compared to manual visual evaluation. Faster image processing rates can be achieved by installing the program on high-end workstations and further improving system performance.
Not only BNCs, but also a large number of MOCs and POCs are produced in the CBMN process. Another benefit of this system is that it can distinguish between cells with differing nucleus numbers. The cell proliferation index40, the ratio of trinuclear cells to tetranuclear cells41, and other information may be calculated using MOC and POC data, which can aid in the estimation of high radiation doses.
Conclusion
This study describes a method for analyzing Giemsa stained CBMN microscopic images quickly and automatically, with a high detection rate and processing speed. This system is simple to use and does not need specifical control over cell density and staining intensity. The BNC and BNCMN detection rates of this system were 99.4% and 89.4%, respectively, of manual detection, while the analysis speed was 40–50 times faster than manual detection. Even when the cells are heavily colored, the method can distinguish BNCs from MNi and MOCs from POCs. As the number of training samples grows, CNN can consistently enhance MN's accuracy and detection rate.
In the future, system testing and analysis will be carried out on samples irradiated with various dosages in order to continuously enhance the system's performance. The software's detection capabilities will also be improved, allowing for the detection of MOCs containing MNi, POCs containing MNi, nucleoplasmic bridges, necrotic cells, and apoptotic cells.
References
Rea, M. E., Gougelet, R. M., Nicolalde, R. J., Geiling, J. A. & Swartz, H. M. Proposed triage categories for large-scale radiation incidents using high-accuracy biodosimetry methods. Health Phys. 98, 136–144 (2010).
IAEA (International Atomic Energy Agency). Cytogenetic Analysis for Radiation Dose Assessment. IAEA Technical Report Series. 405, 14–71 (2001).
Ossetrova, N. I., Sandgren, D. J. & Blakely, W. F. Protein biomarkers for enhancement of radiation does and injury assessment in nonhuman primate total-body irradiation model. Radiat. Prot. Dosimetry. 159, 61–76 (2014).
Rothkamm, K. et al. Comparison of established and emerging biodosimetry assays. Radiat. Res. 180, 111–119 (2013).
Brengues, M. et al. Biodosimetry on small blood volume using gene expression assay. Health Phys. 98, 179–185 (2010).
Turner, H. C. et al. Adapting the gamma-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat. Res. 175, 282–290 (2011).
Sharma, P. M., Ponnaiya, B., Taveras, M., Shuryak, I., M., Turner, H. & Brenner, D.J. High throughput measurement of γH2AX DSB repair kinetics in a healthy human population. PLoS One. https://doi.org/10.1371/journal.pone.0121083 (2015).
Fenech, M. & Morley, A. A. Measurement of micronuclei in lymphocytes. Mutat. Res. Envir. Mut. Rel. Sub. 147, 29–36 (1985).
Repin, M., Pampou, S., Karan, C., Brenner, D. J. & Garty, G. RABiT-II: Implementation of a high-throughput micronucleus biodosimetry assay on commercial biotech robotic systems. Radiat. Res. 187, 492–498 (2017).
Fenech, M. The lymphocyte cytokinesis-block micronucleus cytome assay and its application in radiation biodosimetry. Health Phys. 98, 234–243 (2010).
Senthamizhchelvan, S., Pant, G. S., Rath, G. K., Julka, P. K. & Nair, O. Biodosimetry using micronucleus assay in acute partial body thera-peutic irradiation. Physica Med. 25, 82–87 (2009).
Beinke, C., Port, M., Riecke, A., Ruf, C. G. & Abend, M. Adaption of the cytokinesis-block micronucleus cytome assay for improved triage biodosimetry. Radiat. Res. 185, 461–472 (2016).
Chen, Y. et al. Validating high-throughput micronucleus analysis of peripheral reticulocytes for radiation biodosimetry: benchmark against dicentric and CBMN assays in a mouse model. Health Phys. 98, 218–227 (2010).
Kalantzi, O. I. et al. Low dose induction of micronuclei by lindane. Carcinogenesis 25, 613–622 (2004).
Hong, S. H., Szili, E. J., Fenech, M., Gaur, N. & Short, R. Genotoxicity and cytotoxicity of the plasma jet-treated medium on lymphoblastoid WIL2-NS cell line using the cytokinesis block micronucleus cytome assay. Sci. Rep. 7, 3854 (2017).
Drosopoulou, E. et al. In vitro and in vivo evaluation of the genotoxic and antigenotoxic potential of the major Chios mastic water constituents. Sci. Rep. 8, 12200 (2018).
Doherty, A. T., Hayes, J., Fellows, M., Kirk, S. & O’Donovan, M. A rapid, semi-automated method for scoring micronuclei in mononucleated mouse lymphoma cells. Radiat. Res. 726, 36–41 (2011).
Seager, A. L. et al. Recommendations, evaluation and validation of a semi-automated, fluorescent-based scoring protocol for micronucleus testing in human cells. Mutagenesis 29, 155–164 (2014).
Willems, P. L. et al. Automated micronucleus (MN) scoring for population triage in case of large scale radiation events. Int. J. Radiat. Biol. 86, 2–11 (2010).
Varga, D. et al. An automated scoring procedure for the micronucleus test by image analysis. Mutagenesis 19, 391–397 (2004).
Selyan, G. T., Chaudhury, N. K. & Venkatachalam, R. Comparison of results of the manual and automated scoring of micronucleus frequencies in Co-60-irradiated peripheral blood lymphocytes for triage dosimetry. Appl. Radiat. Isot. 97, 70–77 (2015).
Verhaegen, F. et al. Scoring of radiation-induced micronuclei in cytokinesis-blocked human lymphocytes by automated image analysis. Cytometry 17, 119–127 (1994).
Rodrigues, M. A., Beaton-Green, L. A., Kutzner, B. C. & Wilkins, R. C. Multi-parameter dose estimations in radiation biodosimetry using the automated cytokinesis-block micronucleus assay with imaging flow cytometry. Cytometry A 85, 883–893 (2014).
Rodrigues, M. A., Beaton-Green, L. A., Wilkins, R. C. & Fenech, M. F. The potential for complete automated scoring of the cytokinesis block micronucleus cytome assay using imaging flow cytometry. Mutat. Res. Gene. Tox. Env. Mut. 836, 53–64 (2018).
Wang, Q. et al. Automated triage radiation biodosimetry: integrating imaging flow cytometry with high-throughput robotics to perform the cytokinesis-block micronucleus assay. Radiat. Res. 191, 342–351 (2019).
Repin, M., Pampou, S., Brenner, D. J. & Garty, G. The use of a centrifuge-free RABiT-II system for high-throughput micronucleus analysis. Radiat. Res. 61, 68–72 (2020).
Rodrigues, M. A. An automated method to perform the in vitro micronucleus assay using multispectral imaging flow cytometry. Jove-J. Visua. Exper. 141 (2019).
Wills, J. W. et al. Inter-laboratory automation of the in vitro micronucleus assay using imaging flow cytometry and deep learning. Arch. Toxicol. 95, 3101–3115 (2021).
Rodrigues, M. A. et al. The in vitro micronucleus assay using imaging flow cytometry and deep learning. NPJ Syst. Biol. Appl. 7, 20 (2021).
Decordier, I. et al. Automated image analysis of micronuclei by IMSTAR for biomonitoring. Mutagenesis 26, 163–168 (2011).
Decordier, I. et al. Automated image analysis of cytokinesis-blocked micronuclei: An adapted protocol and a validated scoring procedure for biomonitoring. Mutagenesis 24, 85–93 (2009).
Lyulko, O. V. et al. Fast image analysis for the micronucleus assay in a fully automated high-throughput biodosimetry system. Radiat. Res. 181, 146–161 (2014).
Szirmai, S., Bérces, J. & Köteles, G. J. Computerized image analysis for determining micronucleus frequency. Environ. Health Perspect. 101, 57–60 (1993).
Toossi, M. T. B., Azimian, H., Sarrafzadeh, O., Mohebbi, S. & Soleymanifard, S. Automatic detection of micronuclei by cell microscopic image processing. Mutat. Res. Fund. Mol. Mech. Mut. 806, 9–18 (2017).
Böcker, W., Streffer, C., Müller, W. U. & Yu, C. Automated scoring of micronuclei in binucleated human lymphocytes. Int. J. Radiat. Biol. 70, 529–537 (1996).
Böcker, W., Müller, W. U. & Streffer, C. Image processing algorithms for the automated micronucleus assay in binucleated human lymphocytes. Cytometry 19, 283–294 (1995).
Hulber, T., Kocsis, Z. S., Kis, E., Safrany, G. & Pesznyak, C. A scanning and image processing system with integrated design for automated micronucleus scoring. Int. J. Radiat. Biol. 96, 628–641 (2020).
Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2, 1084–1104 (2007).
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersions simulations. IEEE Trans. Pattern Anal. 13, 583–598 (1991).
Wu, H. et al. Induction of micronuclei in human fibroblasts across the Bragg curve of energetic heavy ions. Radiat. Res. 166, 583–589 (2006).
Müller, W. U. & Rode, A. The micronucleus assay in human lymphocytes after high radiation doses (5–15 Gy). Mutat. Res. Fund. Mol. Mech. Mut. 502, 47–51 (2002).
Acknowledgements
We thank Dr. Yumin Lv and Dr. Lin Han (Henan Institute of Occupational Disease Prevention and Control) for their assistance with sample collection and slide making.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and design of the research. X.S. and Y.C. were instrumental in idea and study design of this research. X.S., C.L. and F.Y. was involved in the programming, data analysis, the main manuscript text. Z.W., J.Z. and Z.Z. were instrumental in manuscript preparation, critical manuscript revision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shen, X., Chen, Y., Li, C. et al. Rapid and automatic detection of micronuclei in binucleated lymphocytes image. Sci Rep 12, 3913 (2022). https://doi.org/10.1038/s41598-022-07936-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-07936-4
This article is cited by
-
Application of image-recognition techniques to automated micronucleus detection in the in vitro micronucleus assay
Genes and Environment (2024)
-
Development of a micronucleus test using the EpiAirway™ organotypic human airway model
Genes and Environment (2023)
-
High-precision automatic identification method for dicentric chromosome images using two-stage convolutional neural network
Scientific Reports (2023)
-
Reply to the opinion paper: is micronucleus test a suitable method for monitoring oral mucosa exposed to dental bleachings in smokers?
Archives of Toxicology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.