Abstract
Aging is associated with widespread physiological changes, including skeletal muscle weakening, neuron system degeneration, hair loss, and skin wrinkling. Previous studies have identified numerous molecular biomarkers involved in these changes, but their regulatory mechanisms and functional repercussions remain elusive. In this study, we conducted next-generation sequencing of DNA methylation and RNA sequencing of blood samples from 51 healthy adults between 20 and 74 years of age and identified aging-related epigenetic and transcriptomic biomarkers. We also identified candidate molecular targets that can reversely regulate the transcriptomic biomarkers of aging by reconstructing a gene regulatory network model and performing signal flow analysis. For validation, we screened public experimental data including gene expression profiles in response to thousands of chemical perturbagens. Despite insufficient data on the binding targets of perturbagens and their modes of action, curcumin, which reversely regulated the biomarkers in the experimental dataset, was found to bind and inhibit JUN, which was identified as a candidate target via signal flow analysis. Collectively, our results demonstrate the utility of a network model for integrative analysis of omics data, which can help elucidate inter-omics regulatory mechanisms and develop therapeutic strategies against aging.
Similar content being viewed by others
Introduction
Aging is a process that accompanies external (physical appearance) and internal (physiological functions) changes in an organism, which are affected by molecular interactions across multiple omics layers. Clinical studies of human aging have identified aging-related biomarkers associated with metabolic disorder1, loss of skeletal muscle2, neurodegeneration3, skin wrinkles4, and hair loss5 as well as an increased risk of aging-related diseases6,7 such as type 2 diabetes, cancers, and Alzheimer’s disease. With the development of next-generation sequencing (NGS) technologies, molecular genetic studies have accumulated large quantities of omics data on aging and aging-related diseases7,8,9.
In particular, blood is easier to collect from living human body than other tissues, and also actively studied in forensic science to estimate the age of suspects10,11. Based on the advantages, numerous association studies on human aging have conducted with blood samples, and revealed aging-related changes in DNA methylation12,13,14,15,16 and gene expression15,16,17,18,19, and their application as biological clocks to estimate chronological age from the identified aging signatures20,21. However, the causal relationship between variations in biomarkers and the specific cellular dysfunction or phenotypes related to aging remains largely unknown. To investigate these molecular mechanisms and causal relationships, molecular dynamics studies have been conducted to predict and control gene expression or protein phosphorylation changes in diseases22,23,24,25,26 such as cancer and diabetes from the perspective of network biology. Several studies have employed ordinary differential equations22,23 or Boolean logical models24,25,26, which require specific parameter fitting or logical rule inference using accumulated experimental data. Recently, a signal flow analysis27,28 method was developed, in which the influence of signals of perturbed nodes on the activity changes of the other nodes can be estimated based on the network structure and mode of action of each edge.
For integrative analysis, investigating the changes in methylation and gene expression in a network model requires understanding the causal relationships between two different omics layers. Changes in DNA methylation states and mRNA expression levels are closely related29. Typically, CpG methylations occurring in TFBSs regulate target gene expression by inhibiting or promoting the binding of specific transcription factors (TFs)30. Furthermore, a decrease in the expression of a gene involved in methylation/demethylation can cause changes in the overall methylation pattern31. It is difficult to determine which methylation/expression event is the leading cause and which is the result of the causal relationship represented in the feedback structures, as DNA methylation and gene expression are regulated by each other.
In this study, we assumed that the gene regulatory network is a closed system and that DNA methylation events are external input signals. Based on this assumption, we investigated the regulation of differentially expressed gene (DEG) markers, which results from changes in the methylation states of differentially methylated position (DMP) markers. First, we conducted paired methyl-seq and RNA-seq analysis of blood samples obtained from 51 Korean individuals aged 21–74 years and identified the DMP and DEG markers associated with age-related changes in methylation states or expression levels. Based on the blood gene regulatory network (GRN) constructed using a tissue-specific network inference study32, we reconstructed an aging-related gene regulatory subnetwork that can explain the changes in gene expression identified via RNA-seq. We further performed signal flow analysis to explore the mechanism by which the methylation states of DMP markers regulate the expression of DEG markers. Furthermore, via signal flow analysis, we identified candidate molecular targets that can reversely regulate the aging-related gene expression changes.
Results
Identification of aging-related DMPs and DEGs
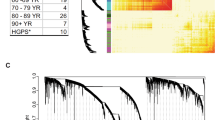
A schematic workflow of multi-dimensional investigation in this study is shown in Fig. 1. NGS analysis was conducted using blood samples obtained from 51 healthy Korean individuals aged 20–74 years. Methyl-seq analysis identified 2,682,537 methylated CpGs from each sample. Of these, the beta values of 222,032 DMPs significantly (P < 0.05) increased or decreased according to age. These DMPs included all 32 aging-related epigenetic markers identified from 100 Korean and 300 Polish samples33 (hypergeometric test, P = 2.3e−35). To use the DMP markers whose methylation states are correlated with expression levels of TF genes as inputs in signal flow analysis of the GRN, we calculated the correlation between the methylation states of each DMP marker and the expression levels of nearby TF genes. The expression levels of 29 TF genes were found to be significantly correlated with the methylation states of 127 DMP markers (P < 0.05). When multiple DMPs were correlated with the same gene, the DMP with the most significant correlation was selected as a representative for calculating the expression level of the gene based on the methylation state of the DMP (Fig. 2c, Supplementary Table S1).
Aging-related DMP and DEG markers identified via NGS. Blood samples were collected from 51 individuals for NGS. (a, b) Heatmap of 22 DMP markers and 46 DEG markers in the aging-related gene regulatory network (GRN). (c) Correlation between the methylation states of DMP markers and the expression levels of neighboring genes. The 22 input DMP markers in the GRN model are indicated as red circles. DEG, differentially expressed gene; DMP, differentially methylated position.
In RNA-seq analysis, we measured the expression levels of 19,229 protein-coding genes in each sample, and found 188 significantly up- or down-regulated DEGs according to age (adjusted P < 0.05; Supplementary Table S2). Of these, 20 DEGs overlapped with 50 aging-related transcriptomic markers identified through a meta-analysis of the blood transcriptome19 (hypergeometric test, P = 8.3e−28).
Reconstruction of an aging-related GRN
To investigate the mechanism underlying how DMP-derived TF expression regulates downstream DEGs through the network, we constructed an aging-related, blood tissue-specific, GRN. Based on a previously constructed blood GRN—comprising 398,283 regulation events between 15,293 genes—in a tissue-specific network inference study32, we reconstructed a subnetwork composed of DMP, DEG markers, and the intermediate nodes via which DMPs regulate downstream DEGs. After filtered out genes and regulations whose expression change or regulatory relationship is not significant based on our RNA-seq experiment, the final aging-related gene regulatory subnetwork was composed of 1,198 regulations among 235 genes (Fig. 3A), including 22 input DMPs (Fig. 2a) and 46 output DEGs (Fig. 2b).
Signal flow analysis of the aging-related GRN constructed with 22 DMP markers and 46 DEG markers. (A) Methylation changes during aging were considered as initial signals for visualization of the aging-related GRN. An increase in edge thickness and node size is based on the intensity of the signal transmitted to the DEG markers via the network. DMP input nodes are indicated in blue; DEG output nodes are indicated in yellow; edges representing up-regulated and down-regulated TG expression are indicated in green and red, respectively. (B) The direction of activity change (DAC) and aging-related fold change in RNA-seq analysis were consistent for the 22 output DEGs. (C) Relative influence of the 22 input DMPs on the DAC of the 22 output DEGs. SFA, signal flow analysis.
Effect of differentially methylated TFs on differentially expressed target genes
To investigate the regulation of the transcriptome layer induced by changes in the epigenomic layer, we performed signal flow analysis, which can predict the direction of activity changes (DAC) of network nodes affected by a given initial signal input. The age-dependent changes in methylation of DMP markers were used as signal input, and their effect on DEG markers via the reconstructed GRN were predicted as DACs. As GRN concerns regulation of gene expression between TFs and their target genes, the initial signal of DMP markers were converted into expression levels of relevant TF genes based on the correlation between DMP marker methylation states and neighboring TF expression levels (Fig. 2c). As a result of signal flow analysis performed on 51 blood samples, the predicted DACs of 26 out of 46 DEGs were significantly altered according to age (P < 0.05). Of these 26 DEGs, the DACs of 22 DEGs were consistent with the increasing/decreasing directions of the DEG markers identified via RNA-seq (Fig. 3B, Supplementary Fig. S1). The remaining four DEGs were inconsistent with the RNA-seq results, although the DAC increased or decreased in a certain direction according to age (Supplementary Figs. S2, S3A). The remaining 20 DEG markers were not compared with the RNA-seq results because their DACs showed nonsignificant correlation with age (P > 0.05; Supplementary Fig. S3B). The 22 DEG markers whose DACs by methylation input were significant and consistent with the RNA-seq results were considered adequately regulated via signal flow analysis of the constructed network model. We accordingly used these markers as targets for reverse regulation to delay or attenuate aging. The relative influence of each DMP in determining the DAC of 22 DEGs was further compared (Fig. 3C).
Candidate molecular targets and drugs to reverse aging-related DEG expression
We investigated candidate molecular targets that reversely regulate the aging-related DEG expression. We performed signal flow analysis assuming up-/down-regulation of each intermediate node as an input signal, and ranked the candidate targets based on the number of DEGs whose DAC are calculated in the opposite direction to its aging-related expression change. We identified top 5% of them as candidate molecular targets (Fig. 4A). Downregulation of GMEB1 or NR3C2 were predicted to reversely regulate 20 of 22 output DEGs, and 15 of them more significantly compared to other input signals. We visualized the calculated DAC of 22 output DEGs for each signal input and the aging-related DEG expression (Fig. 4B).
Candidate target genes identified by signal flow analysis and candidate drugs screened from the LINCS database. (A) List of candidate molecular targets predicted to reversely regulate the aging-related DEG expression. (B) Heatmap of the calculated DACs of the output DEGs for the candidate molecular target. (C) List of candidate drugs identified from the LINCS database that reversely regulate the DEG markers. (D) Heatmap of the observed expression changes of the output DEGs induced by candidate drugs. (E) Subnetwork visualization of candidate targets that reversely regulate the aging-related DEG expression, considering the initial signal that down-regulates JUN (blue). Signals from the candidate target genes (nodes with a black bold boundary) are transmitted via the network with up-regulating (red) or down-regulating (blue) downstream genes, and eventually regulate the aging-related expression of the output DEGs (the color of boundary denotes the expression fold change during aging).
We also investigated candidate drugs that reversely regulate the aging-related expression of the DEGs found in this study, based on a LINCS database34. Of the 12,328 genes with available expression changes in the LINCS database, 19 of the 22 DEG markers found in this study were included. We investigated the drugs and conditions that can reversely regulate these 19 DEGs, and identified 11 candidate drugs, which induced the opposite direction of expression change for more than half of the DEGs (Fig. 4C). We visualized the drug-induced expression change of 19 DEGs and their aging-related expression changes observed in the RNA-seq analysis (Fig. 4D).
Validation of candidate control targets
We investigated the binding targets of the candidate drugs identified with LINCS database to determine whether the drugs regulate the DEG expression by binding to the candidate targets using the PubChem database35,36. We obtained a list of assays in which drug-target binding, and agonist/antagonist experiments were conducted for the LINCS dataset candidate drugs. However, among the eight candidate target genes (Fig. 4A, E), high throughput screening assays were conducted only for NR3C2 and JUN, which can determine target-binding drugs and examine whether they have agonistic or antagonistic action. In the case of NR3C2, 243 assays were conducted for 506 drugs; however, the 11 candidate drugs identified from the LINCS database were not examined in PubChem assays. On the other hand, out of 69 assays where the binding ability of JUN to 7857 drugs was tested, curcumin, which has been found to notably modulate 10 aging-related DEGs in the LINCS database, was observed to bind to JUN and act as an antagonist. Previous studies have shown that administering curcumin increases the life span of model organisms such as mouse37, Drosophila38,39, and Caenorhabditis elegans40; moreover, numerous studies have been conducted on curcumin as a candidate target in aging41,42. For the other candidate genes, PubChem assays were conducted to examine siRNA activity instead of drug compounds, and we could not directly investigate whether drug-target binding occurred. To validate the candidate genes whose drug targets were not directly confirmed due to the lack of drug binding experimental data on the PubChem BioAssay, we further investigated the connection between them and aging via a literature survey; we found changes in aging-related phenotypes following changes in the expression level of relevant genes predicted by signal flow analysis (Table 1).
Discussion
Recent advances in multi-omics integrative analysis have facilitated a detailed understanding of aging-related diseases and the development of therapeutic strategies. Numerous studies have used multi-omics data to estimate chronological age43, identify distinct stages of aging44, investigate interconnectivity of aging-related diseases45, and discover anti-aging drug compounds46. Many of these studies integrated multi-omics data using network approaches; however, there are limitations to analyzing undirected networks based on similarity, conditional dependency, or co-expression as exploring causal relationships among biomolecules is unfeasible. In the present study, we conducted methyl-seq and RNA-seq analysis as well as integrated multi-omics analysis to investigate the regulatory mechanism between methylation and expression according to a GRN.
In network analysis, constructing an accurate network structure is crucial, particularly for signal flow analysis, which mainly relies on the topology information of the network. In general, GRNs that are constructed from databases such as TRANSFAC and STRING can be regarded as canonical networks containing the entire set of gene regulations that can be partially activated or deactivated depending on the conditions. To investigate the GRN in terms of aging, we reconstructed a subnetwork—based on a previously constructed blood-specific GRN—to include genes whose methylation and expression levels changed with age. The observation that TF genes affected by DMP markers modulate downstream genes in a direction consistent with DEG markers demonstrated the existence of a regulatory relationship between the two omics layers.
Based on this regulatory relationship, we identified the candidate control targets (i.e., intermediate nodes) that can reversely regulate the DEG markers. JUN, one candidate target identified via network analysis (Fig. 5A), was validated using the expression profile and binding assay data found in experimental databases. We then found that curcumin, which reversely regulated DEG markers, binds to JUN and inhibits its activity. This indicates that a connection to aging can be discovered by performing additional research or accumulating data on other candidate target genes that have not yet been sufficiently investigated.
Subnetwork visualization. (A) Subnetwork showing the regulation of expression of DEG outputs by DMP inputs; 17 out of 22 DEG output nodes are reversely regulated by JUN inhibition. (B) A subnetwork showing the regulation of output DEGs by JUN inhibition and its downstream regulation. An increase in edge thickness and node size is based on the intensity of the signal transmitted to the DEG markers via the network. Up-regulated and down regulated genes are denoted with red and blue, respectively.
In searching for direct downstream genes of JUN (Fig. 5B), we found that a few downstream genes are regulated in the direction of accelerating aging or promoting aging-related diseases (Table 2). However, whether controlling expression of a specific gene can regulate most downstream genes in the direction of reversing aging does not indicate that all downstream genes are regulated in a consistent direction. Several studies have reported that TF regulation of downstream genes can exert opposing effects on biological processes47 and that a TF can bi-directionally regulate a target gene depending on the cofactor to which it binds48; this is beneficial for achieving homeostasis. Therefore, candidate targets from computational studies should be examined in vitro and in vivo in animals and humans to identify their effects on phenotypes, side effects, and optimal doses in the target tissue.
Finally, regulation of gene expression in this network study only considered part of the epigenome and transcriptome; however, other omics layers involved in modulation before or after transcription, such as chromatin remodeling and protein conformation, can ultimately affect the behavior of biomolecules. Recently, multi-omics data across various other layers have been successfully accumulated on a large scale and with a longitudinal design49,50. When multi-omics network becomes complex and elaborate with the accumulation of data, other network theories can be applied such as reducing the size of biological networks51 or identifying robust control targets that are less affected by timing52. Additionally, clustering methods53,54 developed for time-series profiles can also be applied with the longitudinal data to infer regulatory mechanisms55 or functional annotations56 of molecular markers in each omics layer. Along with advances in theoretical frameworks, employing these qualitative and quantitative multi-omics datasets and phenotype data can improve integrated omics analysis of a network biology and provide new insights that could not be discovered at the level of individual markers or a single omics layer.
Material and methods
Study design and participants
Healthy adult volunteers between 20 and 80 years of age were recruited from two cities in Republic of Korea: Ulsan and Miryang by Welfare Genome Project (WGP)57. Of these, the 51 samples with 24 males (47.1%) and 27 females (52.9%) were sequenced. Their mean age was 44.73 ± 15.02 years. All participants took an empty stomach 9 h before blood collection. The blood samples were frozen at − 80 °C and 15 ml of them were used for Methyl-seq and RNA-seq. Smoking status of participants were collected using a questionnaire, and classified as “smokers” or “non-smokers” (including both former and current smokers). We adjusted for sex bias in identifying DMP and DEG markers, but not for smoking status, since only 8 of 51 participants were smokers and only males (Supplementary Fig. S4).
Ethics, consent, and permissions
All 51 blood samples were obtained from Ulsan University Hospital, and written informed consent was obtained from all participants. This study was approved by the Institutional Review Board of Ulsan National Institute of Science and Technology (approval no. UNISTIRB-16-13-C). All experiments were performed in accordance with relevant guidelines and regulations.
DNA methyl-sequencing
Genomic DNA was isolated from blood using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Extracted DNA was quantified using the Quant-iT BR Assay Kit (Invitrogen, Carlsbad, CA). Genomic libraries were prepared using the SureSelectXT Methyl-Seq Target Enrichment System for Illumina Multiplexed Sequencing (Agilent Technologies, Santa Clara, CA). Briefly, genomic DNA (3 μg per sample) was randomly sheared using an ultrasonicator (Covaris Inc., Woburn, MA), after which DNA fragments were extracted. Samples were then subjected to end repair, methylated adapter ligation, hybridization to SureSelectXT Methyl-Seq Capture Library, streptavidin bead enrichment, bisulfite conversion, and PCR amplification. Sample genomic libraries were indexed and pooled for multiplexed sequencing on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA) using 101 bp paired-end reads.
Read mapping and methylation analysis
The sequenced methyl-seq reads were filtered using the IlluQCPRLL.pl script of NGSQCToolkit (v. 2.3.3)58, and then reads with Q20 > 70% were used. Using Bismark (v.0.14.5)59, the filtered reads were mapped to the human reference genome (hg38; ENSEMBL v.95) and duplicated reads were removed. Methylation values at each site were obtained using MethylExtract (v.1.9.1)60. We filtered out sex chromosome regions to reduce sex bias. The methylation values were normalized per sample using MethylKit package in R (v.3.5.0)61, and adjusted for sex bias using Combat algorithm from SVA package62 in R (v. 3.5.0). Pearson’s correlation coefficient was calculated to identify aging-related DMPs. Multiple test corrections were not applied in the methyl-seq analysis due to the small sample size compared to the large number of CpG positions. To explore the regulatory impact of DMPs on TF expression, we calculated Pearson’s correlation coefficient between DMPs and the expression levels of their neighboring genes of which transcription start site or gene body region are located within 5 kb.
RNA sequencing
Total RNA was extracted using the PAXgene Blood RNA Kit (Qiagen) according to manufacturer’s instructions. RNA quality was assessed using the Bioanalyzer 4200 system to determine the RNA integrity number and rRNA ratio. We purified mRNA from total RNA using polyA selection followed by fragmentation. The fragmented mRNAs were synthesized into single-stranded and double-stranded cDNA using random hexamer priming. RNA sequencing libraries were constructed from double-stranded cDNA using an Illumina TruSeq Stranded mRNA Library Prep Kit. Libraries of the 51 samples were sequenced on the Illumina HiSeq 2500 platform using 101 bp paired-end reads.
Read mapping and expression analysis
The sequenced RNA-seq reads were filtered using the same criteria for methyl-seq analysis with the IlluQCPRLL.pl script of NGSQCToolkit. The filtered reads were aligned to the human genome (hg38; ENSEMBL v.95) by STAR (v. 2.6.0a)63. Gene expression was calculated with the htseq-count tool of the HTSeq software suite (v. 0.11.2)64. DEGs, whose expression varied significantly with age (treated as a continuous variable), were identified with adjusting for sex bias by DESeq2 (v. 1.26.0)65. DESeq2 calculates the log2 fold change of gene expression per unit of age (year) using generalized linear models. Adjusted P value was calculated based on Benjamini–Hochberg correction66.
Blood tissue-specific GRN
To investigate the regulatory relationship between the DMP and DEG markers identified in the blood samples, a blood tissue-specific GRN constructed with the Passing Attributes between Networks for Data Assimilation (PANDA) algorithm67 was used. PANDA is an algorithm that can infer the structure of a GRN from a certain gene expression profile based on previously known TF-target gene binding and protein–protein interaction information. Using 38 tissues in the Genotype-Tissue Expression (GTEx) database68, Sonawane et al.32 constructed tissue-specific GRNs, from which we downloaded the blood tissue-specific GRN (https://zenodo.org/record/838734#.XALkry3MxTZ) and reconstructed an aging-related subnetwork that comprised our identified DMP and DEG markers. Based on our RNA-seq experiment, we refined the subnetwork by filtering out lower 5% genes in their expression variance among 51 blood samples as well as regulations whose correlation and partial correlation of TF-target gene expression were not significant (P > 0.05), so that the refined network structure can better reflect the expression profiles conducted in context of aging.
Determining the direction of gene regulation
In the aging-related GRN constructed based on the GTEx-PANDA network, edges include directional information in terms of source and target; however, they do not include information on the regulatory mode that informs whether a source node promotes or inhibits a target node, which is required to apply signal flow analysis. To explore the regulatory mechanism of gene expression, we assigned the mode of action of each edge according to the correlation of expression levels observed in the RNA-seq experiment of the gene pair to which each edge connects to from the established aging-related GRN. In the aging-related GRN, which has a dense structure of regulatory edges, one target gene was often regulated by multiple TFs. This complex regulatory structure renders the correlation between target gene expression and a certain TF less significant because changes in target gene expression levels via regulation by other TFs acts as noise. In case of a target gene regulated by multiple upstream TF genes, to identify the direction in which a certain TF regulates the target gene, we assigned the mode of action in the direction that corresponds to the partial correlation coefficient69, via which expression changes regulated by other TFs were excluded. Edges with no significant partial correlation were excluded from subsequent analysis.
Signal flow analysis
Signal flow analysis27 is an algorithm that can predict the influence of input signals from specific nodes on the other nodes based on the network structure and the mode of action of each regulation. To use the methylation states of DMP markers as expression signal input given to TF genes in the GRN, the beta values measured in 51 blood samples were normalized as Z-score for each position, and converted into values in a range between − 1 and + 1. DACs of output DEGs were calculated by signal flow analysis. The DEGs whose DAC were concordant with the aging-related expression change in the RNA-seq experiment were considered to be potentially regulated by DMP markers. To determine candidate molecular targets that can reversely regulate the 22 significant DEGs, + 1 and − 1 input signals were assigned to each intermediate node except for the DMP input and DEG output nodes. The DEGs whose DAC were opposite to aging-related change were considered to be controlled in direction to reverse aging. Signal flow analysis was performed with a default parameter (α = 0.5), which adjust the influence of signal flow from upstream genes and the basal activity of their target gene. DACs of output DEGs were calculated as previously described27.
Drug-target profile database
To determine candidate drugs that can reverse DEG marker expression, we used the LINCS database34, which provides the expression level changes of 12,328 genes for a given drug perturbation under different doses and time conditions in various cell lines; expression levels of 978 landmark genes were measured using an array and those of the other genes were inferred from the landmark genes. We downloaded the level 5 modZ score of blood cell-derived B266 cell lines from GEO(GSE92742) data, which included 271 conditions of 47 drug perturbations at different dose and time conditions. We transformed the modZ score of the expression level into percentiles, and counted the number DEGs regulated in the opposite direction to changes with aging with a greater influence than those of the upper/lower tertile (Fig. 3C,D). To compare the capacity for reverse regulation of aging-related DEGs among the different drug compounds, we selected the dosage and time condition that controls the most DEGs in the opposite direction of aging in each drug perturbation.
To validate the candidate targets identified by signal flow analysis, we used PubChem Bioassay35,36, which provide the results of quantitative high-throughput screening assays, wherein a fluorescent target protein reacts with different doses of drug perturbations, and the trend of changes in fluorescence and cell viabilities are measured to examine whether the protein binds to the drug and exhibits an agonistic or antagonistic function.
Data availability
Raw sequencing data of methyl-seq and RNA-seq analysis are freely available upon request and after approval from the Korean Genomics Center’s review board of UNIST. Information regarding data sharing can be found at http://koreangenome.org.
References
Lettieri-Barbato, D., Giovannetti, E. & Aquilano, K. Effects of dietary restriction on adipose mass and biomarkers of healthy aging in human. Aging 8, 3341–3355. https://doi.org/10.18632/aging.101122 (2016).
Yamada, Y. et al. Electrical properties assessed by bioelectrical impedance spectroscopy as biomarkers of age-related loss of skeletal muscle quantity and quality. J. Gerontol. A. 72, 1180–1186. https://doi.org/10.1093/gerona/glw225 (2017).
Parisiadou, L. et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J. Neurosci. 29, 13971–13980. https://doi.org/10.1523/JNEUROSCI.3799-09.2009 (2009).
O’brien, M. E. et al. Loss of skin elasticity is associated with pulmonary emphysema, biomarkers of inflammation, and matrix metalloproteinase activity in smokers. Respir. Res. 20, 128. https://doi.org/10.1186/s12931-019-1098-7 (2019).
Commo, S., Gaillard, O. & Bernard, B. A. Human hair greying is linked to a specific depletion of hair follicle melanocytes affecting both the bulb and the outer root sheath. Br. J. Dermatol. 150, 435–443. https://doi.org/10.1046/j.1365-2133.2004.05787.x (2004).
Gensous, M. et al. The epigenetic landscape of age-related disease: the geroscience perspective. Biogerontology 18, 549–559. https://doi.org/10.1007/s10522-017-9695-7 (2017).
Bacos, K. et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat. Commun. 7, 11089. https://doi.org/10.1038/ncomms11089 (2016).
Teschendorff, A. E. et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 20, 440–446. https://doi.org/10.1101/gr.103606.109 (2010).
Li, H. et al. Identification of molecular alterations in leukocytes from gene expression profiles of peripheral whole blood of Alzheimer’s disease. Sci. Rep. 7, 14027. https://doi.org/10.1038/s41598-017-13700-w (2017).
Anderson, S., Howard, B., Hobbs, G. R. & Bishop, C. P. A method for determining the age of a blood stain. Forensic. Sci. Int. 148, 37–45. https://doi.org/10.1016/j.forsciint.2004.04.071 (2005).
Vidaki, A. et al. DNA methylation-based forensic age prediction using artificial neural networks and next generation sequencing. Forensic. Sci. Int. Genet. 28, 225–236. https://doi.org/10.1016/j.fsigen.2017.02.009 (2017).
Heyn, H. et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 109, 10522–10527. https://doi.org/10.1073/pnas.1120658109 (2012).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. https://doi.org/10.1016/j.molcel.2012.10.016 (2013).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, 3156. https://doi.org/10.1186/gb-2013-14-10-r115 (2013).
Steegenga, W. T. et al. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age 36, 1523–1540. https://doi.org/10.1007/s11357-014-9648-x (2014).
Furukawa, R. et al. Intraindividual dynamics of transcriptome and genome-wide stability of DNA methylation. Sci. Rep. 6, 26424. https://doi.org/10.1038/srep26424 (2016).
Hong, M. G., Myers, A. J., Magnusson, P. K. E. & Prince, J. A. Transcriptome-wide assessment of human brain and lymphocyte senescence. PLoS ONE 3, e3024. https://doi.org/10.1371/journal.pone.0003024 (2008).
Glass, D. et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 14, R75. https://doi.org/10.1186/gb-2013-14-7-r15 (2013).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570. https://doi.org/10.1038/ncomms9570 (2015).
Zhang, Q. et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 11, 54. https://doi.org/10.1186/s13073-019-0667-1 (2019).
Fleischer, J. G. et al. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 19, 221. https://doi.org/10.1186/s13059-018-1599-6 (2018).
Park, S.-M. et al. Systems analysis identifies potential target genes to overcome cetuximab resistance in colorectal cancer cells. FEBS J. 286, 1305–1318. https://doi.org/10.1111/febs.14773 (2019).
Shin, S.-Y. et al. The switching role of β-adrenergic receptor signalling in cell survival or death decision of cardiomyocytes. Nat. Commun. 5, 5777. https://doi.org/10.1038/ncomms6777 (2014).
Park, D. et al. Attractor landscape analysis of the cardiac signaling network reveals mechanism-based therapeutic strategies for heart failure. J. Mol. Cell. Biol. 10, 180–194. https://doi.org/10.1093/jmcb/myj019 (2018).
Choi, M., Shi, J., Zhu, Y., Yang, R. & Cho, K. H. Network dynamics-based cancer panel stratification for systemic prediction of anticancer drug response. Nat. Commun. 8, 1940. https://doi.org/10.1038/s41467-017-02160-5 (2017).
Dutta, P., Ma, L., Ali, Y., Sloot, P. & Zheng, J. Boolean network modeling of β-cell apoptosis and insulin resistance in type 2 diabetes mellitus. BMC Syst. Biol. 13, 36. https://doi.org/10.1186/s12918-019-0692-0 (2019).
Lee, D. & Cho, K. H. Topological estimation of signal flow in complex signaling networks. Sci. Rep. 8, 5262. https://doi.org/10.1038/s41598-018-23643-5 (2018).
Lee, D. & Cho, K. H. Signal flow control of complex signaling networks. Sci. Rep. 9, 14289. https://doi.org/10.1038/s41598-019-50790-0 (2019).
Ozer, B. & Sezerman, U. Analysis of the interplay between methylation and expression reveals its potential role in cancer aetiology. Funct. Integr. Genom. 17, 53–68. https://doi.org/10.1007/s10142-016-0533-9 (2017).
Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356, 2239. https://doi.org/10.1126/science.aaj2239 (2017).
Li, Y., Liu, Y., Strickland, F. M. & Richardson, B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity. Exp. Gerontol. 45, 312–322. https://doi.org/10.1016/j.exger.2009.12.008 (2010).
Sonawane, A. R. et al. Understanding tissue-specific gene regulation. Cell Rep. 21, 1077–1088. https://doi.org/10.1016/j.celrep.2017.10.001 (2017).
Cho, S. et al. Independent validation of DNA-based approaches for age prediction in blood. Forensic Sci. Int. Genet. 29, 250–256. https://doi.org/10.1016/j.fsigen.2017.04.020 (2017).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452. https://doi.org/10.1016/j.cell.2017.10.049 (2017).
Kim, S. et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. https://doi.org/10.1093/nar/gky1033 (2019).
Kim, S., Thiessen, P. A., Cheng, T., Yu, B. & Bolton, E. E. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 46, W563–W570. https://doi.org/10.1093/nar/gky294 (2018).
Kitani, K., Osawa, T. & Yokozawa, T. The effects of tetrahydrocurcumin and green tea polyphenol on the survival of male C57BL/6 mice. Biogerontology 8, 567–573. https://doi.org/10.1007/s10522-007-9100-z (2007).
Lee, K. S. et al. Curcumin extends life span, improves health span, and modulates the expression of age-associated aging genes in Drosophila melanogaster. Rejuv. Res. 13, 561–570. https://doi.org/10.1089/rej.2010.1031 (2010).
Shen, L. R. et al. Curcumin-supplemented diets increase superoxide dismutase activity and mean lifespan in Drosophila. Age 35, 1133–1142. https://doi.org/10.1007/s11357-012-9438-2 (2013).
Liao, V. H. C. et al. Curcumin-mediated lifespan extension in Caenorhabditis elegans. Mech. Aging Dev. 132, 480–487. https://doi.org/10.1016/j.mad.2011.07.008 (2011).
Bielak-Zmijewska, A. et al. The role of curcumin in the modulation of ageing. Int. J. Mol. Sci. 20, 1239. https://doi.org/10.3390/ijms20051239 (2019).
Sikora, E., Bielak-Zmijewska, A., Mosieniak, G. & Piwocka, K. The promise of slow down ageing may come from curcumin. Curr. Pharm. Des. 16, 884–892. https://doi.org/10.2174/138161210790883507 (2010).
Zubakov, D. et al. Human age estimation from blood using mRNA, DNA methylation, DNA rearrangement, and telomere length. Forensic Sci. Int. Genet. 24, 33–43. https://doi.org/10.1016/j.fsigen.2016.05.014 (2016).
Holzscheck, N. et al. Multi-omics network analysis reveals distinct stages in the human aging progression in epidermal tissue. Aging 12, 12393–12409. https://doi.org/10.18632/aging.103499 (2020).
Zierer, J. et al. Exploring the molecular basis of age-related disease comorbidities using a multi-omics graphical model. Sci. Rep. 6, 37646. https://doi.org/10.1038/srep37646 (2016).
Yang, J. et al. Human geroprotector discovery by targeting the converging subnetworks of aging and age-related disease. Geroscience 42, 353–372. https://doi.org/10.1007/s11357-019-00106-x (2020).
Aylon, Y. & Oren, M. The paradox of p53: What, how, and why?. Cold Spring Harb. Perspect. Med. 6, a026328. https://doi.org/10.1101/cshperspect.a026328 (2016).
Chen, D. et al. Bidirectional regulation of osteogenic differentiation by the FOXO subfamily of Forkhead transcription factors in mammalian MSCs. Cell Prolif. 52, e12540. https://doi.org/10.1111/cpr.12540 (2018).
De Jager, P. L. et al. Data Descriptor: A multi-omics atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142. https://doi.org/10.1038/sdata.2018.142 (2018).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26, 83–90. https://doi.org/10.1038/s41591-019-0719-5 (2020).
Kim, J. R. et al. Reduction of complex signaling networks to a representative kernel. Sci. Signal 4, 35. https://doi.org/10.1126/scisignal.2001390 (2011).
Park, S. J. & Cho, K. H. Delay-robust supervisory control of discrete-event systems with bounded communication delays. IEEE Trans. Automat. Contr. 51, 911–915. https://doi.org/10.1109/TAC.2006.872834 (2006).
Möller-Levet, C. S., Cho, K. H. & Wolkenhauer, O. DNA microarray data clustering based on temporal variation: Fcv with tsd preclustering. Appl. Bioinform. 2, 35–45 (2003).
Subhani, N., Rueda, L., Ngom, A. & Burden, C. J. Multiple gene expression profile alignment for microarray time-series data clustering. Bioinformatics 26, 2281–2288. https://doi.org/10.1093/bioinformatics/btq422 (2010).
Meng, J., Gao, S. J. & Huang, Y. Enrichment constrained time-dependent clustering analysis for finding meaningful temporal transcription modules. Bioinformatics 25, 1521–1527. https://doi.org/10.1093/bioinformatics/btp235 (2009).
Tchagang, A. B., Bui, K. V., McGinnis, T. & Benos, P. V. Extracting biologically significant patterns from short time series gene expression data. BMC Bioinform. 10, 255. https://doi.org/10.1186/1471-2105-10-255 (2009).
Jeon, Y. et al. Welfare genome project: A participatory Korean personal genome project with free health check-up and genetic report followed by counseling. Front. Genet. 12, 633731. https://doi.org/10.3389/fgene.2021.633731 (2021).
Patel, R. K. & Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619. https://doi.org/10.1371/journal.pone.0030619 (2012).
Krueger, F. & Andrews, S. R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. https://doi.org/10.1093/bioinformatics/btr167 (2011).
Barturen, G., Rueda, A., Oliver, J. L. & Hackenberg, M. MethylExtract: High-quality methylation maps and SNV calling from whole genome bisulfite sequencing data. F1000 Res. 2, 217. https://doi.org/10.12688/f1000research.2-217.v2 (2013).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87. https://doi.org/10.1186/gb-2012-13-10-R87 (2012).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. https://doi.org/10.1093/bioinformatics/bts034 (2012).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. https://doi.org/10.1093/bioinformatics/btu638 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x (1995).
Glass, K., Huttenhower, C., Quackenbush, J. & Yuan, G. C. Passing messages between biological networks to refine predicted interactions. PLoS ONE 8, e6483210. https://doi.org/10.1371/journal.pone.0064832 (2013).
Lonsdale, J. et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585. https://doi.org/10.1038/ng.2653 (2013).
Porth, I., White, R., Jaquish, B. & Ritland, K. Partial correlation analysis of transcriptomes helps detangle the growth and defense network in spruce. New Phytol. 218, 1349–1359. https://doi.org/10.1111/nph.15075 (2018).
An, W. et al. Glucocorticoid modulatory element-binding protein 1 (GMEB1) interacts with the de-ubiquitinase USP40 to stabilize CFLARL and inhibit apoptosis in human non small cell lung cancer cells. J. Exp. Clin. Cancer Res. 38, 181. https://doi.org/10.1186/s13046-019-1182-3 (2019).
Jung, Y. et al. An epigenetic mechanism mediates developmental nicotine effects on neuronal structure and behavior. Nat. Neurosci. 19, 905–914. https://doi.org/10.1038/nn.4315 (2020).
DuPont, J. et al. Vascular mineralocorticoid receptor regulates microRNA-155 to promote vasoconstriction and rising blood pressure with aging. JCI Insight 8, e88942. https://doi.org/10.1172/jci.insight.88942 (2016).
Bigas, J., Sevilla, L. M. & Pérez, P. Epidermal mineralocorticoid receptor inactivation affects the homeostasis of all skin layers in chronologically aged mice. J. Invest. Dermatol. 127, 514–525. https://doi.org/10.1016/j.jid.2020.03.933 (2020).
Bortell, N. et al. Sirtuin 1-chromatin-binding dynamics points to a common mechanism regulating inflammatory targets in SIV infection and in the aging brain. J. Neuroimmune Pharmacol. 13, 163–178. https://doi.org/10.1007/s11481-017-9772-3 (2018).
Zhou, H., Beevers, C. S. & Huang, S. Targets of curcumin. Curr. Drug Targets 12, 332–347. https://doi.org/10.2174/138945011794815356 (2011).
Dong, J. et al. miR-10a rejuvenates aged human mesenchymal stem cells and improves heart function after myocardial infarction through KLF4. Stem Cell Res. Ther. 9, 151. https://doi.org/10.1186/s13287-018-0895-0 (2018).
Li, J. et al. miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4. J. Cell. Physiol. 228, 2324–2336. https://doi.org/10.1002/jcp.24402 (2013).
Tomczyk, S. et al. Loss of neurogenesis in aging hydra. Dev. Neurobiol. 79, 479–796. https://doi.org/10.1002/dneu.22676 (2017).
Haibo, W. et al. Retinoic acid-induced upregulation of miR-219 promotes the differentiation of embrionic stem cells into neural cells. Cell Death Dis. 8, e2953. https://doi.org/10.1038/10.1038/cddis2017.336 (2020).
Tharyan, R. G. et al. NFYB-1 regulates mitochondrial function and longevity via lysosomal prosaposin. Nat. Metab. 2, 387–396. https://doi.org/10.1038/s42255-020-0200-2 (2020).
Kelly, J., Moyeed, R., Carroll, C., Luo, S. & Li, X. Genetic networks in Parkinson’s and Alzheimer’s disease. Aging 12, 5221–5243. https://doi.org/10.18632/aging.102943 (2020).
Xue, X., Zhang, J., Lan, H., Xu, Y. & Wang, H. Kaiso protects human umbilical vein endothelial cells against apoptosis by differentially regulating the expression of B-cell CLL/lymphoma 2 family members. Sci. Rep. 7, 7116. https://doi.org/10.1038/s41598-017-07559-0 (2017).
Zhenilo, et al. DeSUMOylation switches Kaiso from activator to repressor upon hyperosmotic stress. Cell Death Differ. 25, 1938–1951. https://doi.org/10.1038/s41418-018-0078-7 (2018).
Yi, Y. et al. Ultraviolet A irradiation induces senescence in human dermal fibroblasts by down-regulating DNMT1 via ZEB1. Aging 10, 212–228. https://doi.org/10.18632/aging.101383 (2018).
Ninfali, C., Siles, L., Darling, D. S. & Postigo, A. Regulation of muscle atrophy-related genes by the opposing transcriptional activities of ZEB1/CtBP and FOXO3. Nucleic Acids Res. 46, 10697–10708. https://doi.org/10.1093/nar/gky835 (2018).
Manocha, G. D. et al. NFATc2 modulates microglial activation in the AβPP/PS1 mouse model of Alzheimer’s disease. J. Alzheimers Dis. 58, 775–787. https://doi.org/10.3233/JAD-151203 (2017).
Sarikhani, M. et al. SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J. Biol. Chem. 293, 5281–5294. https://doi.org/10.1074/jbc.RA117.000915 (2018).
Mao, G. X. et al. Salidroside protects human fibroblast cells from premature senescence induced by H2O2 partly through modulating oxidative status. Mech. Ageing Dev. 131, 723–731. https://doi.org/10.1016/j.mad.2010.10.003 (2010).
Mao, G. X. et al. Protective role of salidroside against aging in a mouse model induced by D-galactose. Biomed. Environ. Sci. 23, 161–166. https://doi.org/10.1016/50895-3988(10)60047-5 (2010).
Gao, J. et al. Salidroside ameliorates cognitive impairment in a D-galactose-induced rat model of Alzheimer’s disease. Behav. Brain Res. 293, 27–33. https://doi.org/10.1016/j.bbr.2015.06.045 (2015).
Wang, J. et al. The effect of synthetic salidroside on cytokines and airway inflammation of asthma induced by diisocyanate (TDI) in mice by regulating GATA3/T-bet. Inflammation 38, 697–704. https://doi.org/10.1007/s10753-014-9979-x (2015).
Rafatian, G. et al. Mybl2 rejuvenates heart explant-derived cells from aged donors after myocardial infarction. Aging Cell 19, e13174. https://doi.org/10.1111/acel.13174 (2020).
Avelar, R. A. et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 21, 91. https://doi.org/10.11186/s13059-020-01990-9 (2020).
Deng, Y.-N., Xia, Z., Zhang, P., Ejaz, S. & Liang, S. Transcription factor RREB1: From target genes towards biological functions. Int. J. Biol. Sci. 16, 1463–1473. https://doi.org/10.7150/ijbs.40834 (2020).
Vargas, D. M., De Bastiani, M. A., Zimmer, E. R. & Klamt, F. Alzheimer’s disease master regulators analysis: Search for potential molecular targets and drug repositioning candidates. Alzheimers Res. Ther. 10, 59. https://doi.org/10.1186/s13195-018-0394-7 (2018).
Muñoz, P., Blanco, R., Flores, J. M. & Blasco, M. A. PF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 37, 1063–1071. https://doi.org/10.1038/ng.1633 (2005).
Richter, T. et al. TRF2 overexpression diminishes repair of telomeric single-strand breaks and accelerates telomere shortening in human fibroblasts. Mech. Aging Dev. 128, 340–345. https://doi.org/10.1016/j.mad.2007.02.003 (2007).
Kim, G. et al. The heat shock transcription factor Hsf1 is downregulated in DNA damage–associated senescence, contributing to the maintenance of senescence phenotype. Aging Cell 11, 617–627. https://doi.org/10.1111/j.1474-9726.2012.00827.x (2012).
Cigliano, A. et al. Inhibition of HSF1 suppresses the growth of hepatocarcinoma cell lines in vitro and AKT-driven hepatocarcinogenesis in mice. Oncotarget 8, 54149–54159. https://doi.org/10.18632/oncotarget.16927 (2017).
Chen, Y.-F. et al. Targeting HSF1 leads to an antitumor effect in human epithelial ovarian cancer. Int. J. Mol. Med. 39, 1564–1570. https://doi.org/10.3892/ijmm.2017.2978 (2017).
Luo, R. et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 16, 52–69. https://doi.org/10.1080/15548627.2019.1596488 (2020).
Nogueira-Recalde, U. et al. Fibrates as drugs with senolytic and autophagic activity forosteoarthritis therapy. EBioMedicine 45, 588–605. https://doi.org/10.1016/j.ebiom.2019.06.049 (2019).
Uysal, F. & Ozturk, S. The loss of global DNA methylation due to decreased DNMT expression in the postnatal mouse ovaries may associate with infertility emerging during ovarian aging. Histochem. Cell. Biol. 154, 301–314. https://doi.org/10.1007/s00418-020-01890-w (2020).
Yajima, H. et al. Six family genes control the proliferation and differentiation of muscle satellite cells. Exp. Cell Res. 316, 2932–2944. https://doi.org/10.1016/j.yexcr.2010.08.001 (2010).
Yajima, H. & Kawakami, K. Low Six4 and Six5 gene dosage improves dystrophic phenotype and prolongs life span of mdx mice. Dev. Growth Differ. 58, 546–561. https://doi.org/10.1111/dgd.12290 (2016).
Roussel-Gervais, A. et al. Loss of the methyl-CpG-binding protein ZBTB4 alters mitotic checkpoint, increases aneuploidy, and promotes tumorigenesis. Cancer Res. 77, 62–73. https://doi.org/10.1158/0008-5472.CAN-16-1181 (2017).
Weber, A. et al. Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J. 27, 1563–1574. https://doi.org/10.1038/emboj.2008.85 (2008).
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korean Government the Ministry of Science and ICT (Grant No. 2020R1A2B5B03094920), the KAIST Grand Challenge 30 Project, and the Electronics and Telecommunications Research Institute (ETRI) funded by the Korean government (Grant No. 21ZS1100; Core Technology Research for Self-Improving Integrated Artificial Intelligence System). This work was also supported by the Leading Business for Happy Living for Ulsan and Miryang (Genome-based Biomedical Industry Promotion Business) funded by the Ulsan City Research Fund and Miryang City Research Fund (Grant Nos. 2.160475.01, 2.170010.01, and 2.180016.01); the Research Project funded by the Ulsan City Research Fund (Grant No. 1.200047.01) of Ulsan National Institute of Science & Technology (UNIST); and the U-K Brand Research Fund (GRANT NO. 1.200108.01) of UNIST. Human-derived materials used in this study were obtained from the BioBank of Ulsan University Hospital (60SA2016001-001 and 60SA2016001-003). We appreciate all participants and Ulsan as well as Miryang citizens who supported the Leading Business for Happy Living for Ulsan and Miryang project (Genome-based Biomedical Industry Promotion Business).
Author information
Authors and Affiliations
Contributions
K.-H.C. conceived and supervised the study; J.B. and Y.S.C. planned the experiments; Y.K.K. performed wet-laboratory experiments including DNA and RNA extraction; Y.J., J.Y.J., and H.-Y.L. performed NGS analysis; H.-Y.L. curated the data and performed network analysis; H.-Y.L., Y.J., and K.-H.C. discussed and contributed to the interpretation of the results; and H.-Y.L. and K.-H.C. wrote the manuscript. All authors discussed and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, HY., Jeon, Y., Kim, Y.K. et al. Identifying molecular targets for reverse aging using integrated network analysis of transcriptomic and epigenomic changes during aging. Sci Rep 11, 12317 (2021). https://doi.org/10.1038/s41598-021-91811-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-91811-1
This article is cited by
-
The Information Theory of Aging
Nature Aging (2023)
-
NETISCE: a network-based tool for cell fate reprogramming
npj Systems Biology and Applications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.