Abstract
SHANK2 mutations have been identified in individuals with neurodevelopmental disorders, including intellectual disability and autism spectrum disorders (ASD). Using CRISPR/Cas9 genome editing, we obtained SH-SY5Y cell lines with frameshift mutations on one or both SHANK2 alleles. We investigated the effects of the different SHANK2 mutations on cell morphology, cell proliferation and differentiation potential during early neuronal differentiation. All mutant cell lines showed impaired neuronal differentiation marker expression. Cells with bi-allelic SHANK2 mutations revealed diminished apoptosis and increased proliferation, as well as decreased neurite outgrowth during early neuronal differentiation. Bi-allelic SHANK2 mutations resulted in an increase in p-AKT levels, suggesting that SHANK2 mutations impair downstream signaling of tyrosine kinase receptors. Additionally, cells with bi-allelic SHANK2 mutations had lower amyloid precursor protein (APP) expression compared to controls, suggesting a molecular link between SHANK2 and APP. Together, we can show that frameshift mutations on one or both SHANK2 alleles lead to an alteration of neuronal differentiation in SH-SY5Y cells, characterized by changes in cell growth and pre- and postsynaptic protein expression. We also provide first evidence that downstream signaling of tyrosine kinase receptors and amyloid precursor protein expression are affected.
Similar content being viewed by others
Introduction
Neurodevelopmental disorders have a high prevalence (1–3%) and are often caused by genetic factors. Genes of the SHANK (SH3 and multiple ankyrin repeat domains protein) family (comprising SHANK1, SHANK2 and SHANK3) have been linked to a spectrum of neurodevelopmental disorders1. Deleterious de novo SHANK2 mutations have been identified in individuals with intellectual disability (ID), autism spectrum disorders (ASD), developmental delay, and attention deficit and hyperactivity disorder2,3,4,5. In the SFARI gene database6 SHANK2 is categorized as a high confidence autism risk gene. In addition, SHANK2 has been linked to the pathology of neuropsychiatric (schizophrenia, bipolar disorder)7,8,9,10 and neurodegenerative disorders11.
SHANK2 encodes for a postsynaptic scaffolding protein at glutamatergic synapses in the brain, essential for proper synapse formation, development and plasticity12,13. As organizer of a large protein complex, the so-called postsynaptic density, it connects the different types of glutamate receptors (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA), N-methyl-d-aspartat receptor (NMDA), metabotropic glutamate receptors) to molecules of many signaling pathways and to the actin cytoskeleton14. Impaired insulin signaling in the brain was postulated to contribute to the development of autism in genetically susceptible individuals15. As SHANK2 directly interacts with IRSp53 (insulin receptor substrate p53)16,17, it may be involved in insulin signaling in the brain, making this pathway susceptible to SHANK2 mutations. In addition, studies in mice and human iPSC-derived neurons provided first evidence that a loss of SHANK could be partially rescued by administration of insulin like growth factor 118,19,20. Moreover, SHANK dysregulation may also contribute to the molecular pathology of Alzheimer´s disease (AD)21. In synaptosomes isolated from the middle frontal gyrus from patients with AD increased SHANK2 levels were reported11. Mutations in the gene coding for amyloid precursor protein (APP) have been identified in several individuals with AD22,23. Both SHANK2 and APP are expressed during neuronal differentiation and are involved in synapse formation and neural plasticity23,24,25, and thus could potentially interact in the course of ASD and neurodegenerative disorders.
SHANK2 gene variants have first been investigated in primary hippocampal cultures of rodents by overexpressing the respective mutated protein5,26. These analyses point to an impaired signal transduction at glutamatergic synapses, with a reduced synaptic density at dendrites5,7 and reduced AMPA receptors at the cell surface26, which are impairments that potentially underlie different neurodevelopmental disorders. Shank2 knockout mice and rats have been generated, which show hyperactivity, repetitive behavior as well as social and cognitive impairments together with impaired synaptic transmission27,28,29,30. Recently, ASD-related SHANK2 mutations were investigated in induced pluripotent stem cell-derived cortical neurons, reporting increased dendritic length and complexity as well as a perturbation of the expression of neurodevelopmental genes31. Further information about the functional impact of SHANK2 mutations in humans is needed for a better understanding of the molecular pathology of neurodevelopmental disorders.
In this study we employed the human neuroblastoma cell line SH-SY5Y to model SHANK2 variants. This cell line is a frequently cited in vitro model in neuropsychiatric research. Despite its tumor origin, its neuro-ectodermal lineage allows the investigation of neuronal phenotypes in the context of neurodevelopmental and neurodegenerative diseases32. SH-SY5Y cells express all three members of the SHANK gene family (SHANK1, SHANK2, SHANK3) and have been used to investigate the influence of sex hormones on SHANK expression33. Furthermore, the cells can be differentiated into neurons to model early neurodevelopmental processes of cortical development32, which is relevant as cortical function is central in ASD pathology34,35. Findings in this cellular model nevertheless should also be reproduced in a more physiological model, in order to strengthen the respective findings.
The aim of this study was to elucidate the impact of mono- and bi-allelic SHANK2 frameshift mutations on different cellular properties during early neuronal differentiation by analyzing cell proliferation, neurite morphology, as well as neuronal and glutamatergic marker gene/protein expression. In addition, we explored the effect on receptor tyrosine kinase downstream signaling and on amyloid precursor protein (APP) expression to further improve our understanding of SHANK2 function.
Results
Genome editing of SH-SY5Y cells to generate SHANK2 mutant lines
To investigate the functional consequences of SHANK2 mutations that have been identified in patients with ASD and intellectual disability, we introduced SHANK2 frameshift mutations in SH-SY5Y cells using CRISPR/Cas9-based genome editing. A guide RNA was designed to target the genomic region adjacent to the position of the SHANK2-R841X variant, which had previously been identified in a patient with ASD4. The non-homologous end-joining repair mechanism of the cells led to the introduction of a heterozygous adenosine insertion, which caused a frameshift and premature stop codon (6H6, p.L837Sfs*49). Additionally, we generated a compound heterozygous mutation line (6A4) with an adenosine deletion on one allele and an adenosine insertion on the other, resulting in two different premature stop codons (p.L837Sfs*49, p.L837Wfs*14) (Table 1). As a control, we selected cells that underwent the complete process of genome-editing without being edited (1F11) as well as the original SH-SY5Y line (WT). Mutations were confirmed after several passages (Supplementary Fig. 1) and putative predicted off-target sites were ruled out by Sanger sequencing of the respective genomic regions (Supplementary Table 1).
Western blot experiments revealed a complete loss of SHANK2 wildtype expression for the compound heterozygous cell line and a reduction of SHANK2 wildtype protein by one half for the heterozygous SHANK2 mutant lines (Fig. 1, Supplementary Fig. 2). The SHANK2 antibody targets amino acids 849–1029, a region downstream of the introduced frameshift mutation (6A4: p.L837Sfs*49, p.L837Wfs*14, 6H6:p.L837Sfs*49), therefore the reduction in protein levels reflects the diminished amount of wildtype SHANK2 protein, whereas the presence of a truncated version of the protein cannot be excluded. Treatment with cycloheximide, which blocks nonsense-mediated mRNA decay (NMD), did not reveal any evidence for NMD in both cell lines (Supplementary Fig. 3) even though quantitative real-time PCR (qPCR) analysis of SHANK2 mRNA expression revealed significantly reduced levels in the cell line with compound heterozygous SHANK2 mutations (Supplementary Fig. 4A).
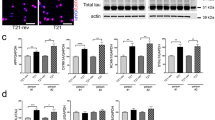
Western blot analysis of SHANK2 expression in genome-edited cell lines (6A4, 6H6) versus the non-edited controls (1F11, WT). SHANK2-expression was normalized against the expression of GAPDH (reference) in undifferentiated cells (UN) and in cells after 7 days of neuronal differentiation (DIF7). Cropped western blot membrane images are shown and full-length blots are presented in Supplementary Fig. 2. n = 4 experiments. One-way ANOVA was performed and the two different mutant cell lines as well as the WT control were compared to the control 1F11 (ANOVA results: SHANK2-UN F = 266.8, P < 0.0001; SHANK2-DIF7 F = 229.8, P < 0.0001). Correction for multiple testing with Dunnett’s test (n = 3 tests for each time point). Data were presented as box-plots and the corrected P-values are indicated. ***P ≤ 0.001.
Cells with bi-allelic SHANK2 mutations show increased proliferation and lower apoptosis rates compared to controls during neuronal differentiation
To assess basic properties of the SHANK2-mutant cell lines, we analyzed cell proliferation at the undifferentiated stage. Immunofluorescence microscopy with Ki67, a cell proliferation marker, showed similar proliferation rates for all lines. Apoptosis was analyzed with the TUNEL assay, which revealed low apoptosis rates under basic conditions without any differences between the cell lines (Fig. 2).
Analysis of apoptosis and cell proliferation in SHANK2 mutant cell lines. A TUNEL assay was carried out to determine the rate of apoptotic cells and TUNEL-positive cells were quantified relative to the total number of nuclei (Hoechst), revealing significantly lower numbers of TUNEL-positive cells for both SHANK2 mutant lines compared to the control 1F11 (one-way ANOVA, F = 22.74, P < 0.0001, P-values after correction for multiple testing with Dunnett’s test 6A4 P < 0.0001, 6H6 P = 0.0004, WT P = 0.326) after 7 days of differentiation (DIF7). In addition, immunofluorescence microscopy was carried out with the proliferation marker Ki67 followed by quantification of the Ki67 positive cells versus the total number of nuclei. A significantly increased number of Ki67-positive cells was identified in cell line 6A4 with bi-allelic SHANK2 mutation in comparison to the control 1F11 (one-way ANOVA, F = 20.97, P < 0.0001, P-values after correction for multiple testing with Dunnett´s test 6A4 P < 0.0001, 6H6 P = 0.49, WT P = 0.327) at DIF7. The pictures illustrate the expression after 7 days of differentiation. No difference was observed between the two control lines WT and 1F11. Undifferentiated cell lines showed equally low apoptosis levels (one-way ANOVA, F = 2.285, P = 0.09, corrected P-values Dunnett´s test 6A4 P = 0.078, 6H6 P = 0.98, WT P = 0.99) and equal levels of cell proliferation (one-way ANOVA, F = 1.12, P = 0.3478, corrected P-values Dunnett´s test 6A4 P = 0.30, 6H6 P = 0.99, WT P = 0.40). Data were presented in box plots. n = 3 experiments. ***P ≤ 0.001.
Next, we differentiated the SH-SY5Y lines according to an improved neuronal differentiation protocol published by Chiocchetti et al.32. With this protocol, SH-SY5Y cells can be differentiated into neurons that express genes relevant in early cortical development and associated to neurodevelopmental/-psychiatric disorders32. To validate the process of neuronal differentiation, we quantified the expression of selected marker proteins by Western blot analysis in the control cell line (WT) under basic culturing conditions and after 7 days of differentiation. A significant increase of the neuronal marker β3-Tubulin (TUBB3) together with decreased levels of the neuronal stem cell protein Nestin (NES) after 7 days of differentiation indicate that the SH-SY5Y cells are starting to differentiate into neurons (Supplementary Figs. 5 and 6). This finding is also supported by an increased expression of the presynaptic marker protein Synaptophysin (SYP) and the postsynaptic density protein 95 (PSD-95) (Supplementary Figs. 5 and 6). Expression analysis of AKT Serine/Threonine Kinase 1 (AKT) was also included, as it is an important mediator of growth factor-induced neuronal survival in the developing nervous system. No significant expression differences could be found for AKT, SHANK2 and the ionotropic glutamate receptor NMDA type subunit 2B (GRIN2B) (Supplementary Figs. 5 and 6).
As we could detect elevated protein levels of glutamatergic (SYP, PSD-95) and pan-neuronal (TUBB3) proteins in parallel to the reduction of the neuronal stem cell marker Nestin after 7 days of differentiation, we selected this stage for further analysis to investigate impairments in early neuronal differentiation. We analyzed cell proliferation after 7 days of differentiation when wildtype cells still proliferate with low apoptosis rates. The cell line 6A4 with bi-allelic SHANK2 mutation showed significantly more Ki67-positive cells, indicating a higher rate of cell proliferation compared to controls, whereas the heterozygous line 6H6 behaved similarly to the controls (Fig. 2). However, reduced numbers of TUNEL-positive cells were identified for both cell lines with SHANK2 mutations (6A4 and 6H6), indicating a lower apoptosis rate compared to controls (Fig. 2).
Bi-allelic SHANK2 mutation impairs neurite outgrowth
To identify morphological impairments in early neuronal differentiation processes, we analyzed neurite outgrowth after 4 days of differentiation in the different cell lines by measuring the length of β3-Tubulin-positive cellular extensions in FiJi36. The control cells (WT, 1F11) extended long neurites and started to form neuronal networks (Fig. 3). The heterozygous SHANK2 mutant line 6H6 developed similarly to the controls, whereas the compound heterozygous SHANK2 mutant line (6A4) showed an obvious impairment regarding network formation and neurite extension. The 6A4 cell line had significantly shorter neurites compared to controls and 6H6 (Fig. 3).
Analysis of cell morphology. Immunofluorescence microscopy was carried out with β3-Tubulin and the length of neurites after 4 days of differentiation was measured, revealing significantly shorter neurites for the cell line with compound heterozygous SHANK2 mutation (6A4) in comparison to the control 1F11. n = 3 experiments, one-way ANOVA (F = 31.1, P < 0.0001), corrected P-values from Dunnett’s test (6A4 P < 0.0001, 6H6 P = 0.36, WT P = 0.48). No difference was observed between the two control lines WT and 1F11. Data were presented in box plots. ***P ≤ 0.001.
Changes of gene expression in SHANK2 mutant cells
To further elucidate the influence of SHANK2 mutations on basic cellular and neuronal differentiation properties, we analyzed mRNA expression levels of several genes involved in these processes by qPCR (n = 3 experiments) (Supplementary Fig. 4). The bi-allelic mutation of SHANK2 in SH-SY5Y cells initially led to strongly reduced SHANK2 mRNA levels compared to the control in the undifferentiated state. After 7 days of differentiation, however, SHANK2 mRNA levels were equal between the cell lines and after 16 days of differentiation even an increase of SHANK2 mRNA expression was found for the compound heterozygous cell line (Supplementary Fig. 4). We also analyzed the gene expression of microtubule-associated protein 2 (MAP2), the presynaptic gene synaptophysin (SYP) and the postsynaptic gene discs large MAGUK scaffold protein 4 (DLG4, encoding for PSD-95) to determine if neuronal, presynaptic and postsynaptic genes are differentially expressed. Furthermore, the expression of all SHANK family members (SHANK1, SHANK2, SHANK3) was analyzed as well as several glutamate receptor genes, as SHANKs are known to interact with glutamate receptors: AMPA receptor subunit 2 (GRIA2), metabotropic glutamate receptor 1 (GRM1) and the ionotropic glutamate receptor kainate type 2 (GRIK2). To find out if GABAergic neurons or aminergic neurons are affected we included GABA receptor beta 3 (GABRB3) and thyroxin hydroxylase (TH) expression analysis. The neuronal stem cell gene Nestin (NES) has been included to determine the presence of very immature neuronal cells.
In undifferentiated cells, a significantly increased expression of NES and a nominally significant reduction in the expression of both SHANK1 and SHANK2 was observed in the compound heterozygous line 6A4 (Supplementary Fig. 4A). Furthermore, expression of GRIA2 and GABRB3 were reduced. A gene predominantly expressed in aminergic neurons, tyrosine hydroxylase, was expressed at low levels with slightly elevated levels in the heterozygous cell line 6H6. Nestin levels remained robustly elevated in the cell line with a bi-allelic SHANK2 mutation (6A4) from the undifferentiated state throughout neuronal differentiation after 7 and 16 days (Supplementary Fig. 4A–C). The results reached statistical significance after correction for multiple testing. Additionally, we found reduced levels of tyrosine hydroxylase in the compound heterozygous cell line (6A4) after 7 and 16 days of differentiation (Supplementary Fig. 4B,C).
After 16 days of differentiation, increased expression levels of the presynaptic gene SYP, all three members of the SHANK family as well as GRIA2 and GABRB3 were identified in the compound heterozygous SHANK2 mutant line (Supplementary Fig. 4). No mRNA expression differences in SHANK2 mutant lines were detected, neither for DLG4, GRM1, GRIK2, nor MAP2 (Supplementary Fig. 4C).
Changes of protein expression in SHANK2 mutant cells
Next, we assessed the expression of several proteins by Western blot to elucidate the consequences of the loss of SHANK2 regarding neuronal differentiation. SHANK2 expression was reduced in the undifferentiated state and after 7 days of differentiation in both SHANK2 mutant lines (6A4, 6H6) compared to the control 1F11 (Fig. 1). We included the non-edited wildtype SH-SY5Y cells (WT) in the Western blot analysis as a second control cell line, and compared expression levels to 1F11. The comparison between the two control lines revealed similar expression levels for WT and 1F11 for all analyzed proteins (Fig. 4). A comparable expression of all tested genes was also shown on mRNA level (Supplementary Fig. 7).
Quantification of protein expression in SH-SY5Y cells (7 days after differentiation). Western blot analysis with protein from two control and two SHANK2 mutant cell lines. The protein expression levels of NESTIN (NES), GRIN2B, PSD95, AKT, β3-Tubulin (TUBB3) and SYP were normalized against the expression of GAPDH (reference). GAPDH expression was analyzed against total protein expression and a differential expression of GAPDH between the different cell lines was ruled out. No differences between the two control lines WT and 1F11 could be detected. Differential expression levels between mutant and control cell lines were found for NES, PSD95 and SYP. Cropped western blot membrane images are shown and full-length blots are presented in Supplementary Fig. 8. n = 5 experiments, one-way ANOVA was carried out and protein expression of the two mutant cell lines (6A4, 6H6) and the wildtype cell line (WT) was compared against the control 1F11 (ANOVA results: NES F = 11.57, P = 0.0007; GRIN2B F = 1.21, P = 0.348; PSD95 F = 63,51, P < 0.0001; AKT F = 1.91, P = 0.181; TUBB3 F = 0.28, P = 0.837; SYP F = 6.48, P = 0.0074; GAPDH F = 0.93, P = 0.458). n.s.—not significant. Correction for multiple testing with Dunnett´s test (n = 3 tests for each protein). Data were presented as box-plots and the corrected P-values are indicated, *P ≤ 0.05, ***P ≤ 0.001.
To elucidate the influence of SHANK2 mutations on differentiated cells, we compared protein expression levels between the control 1F11 and the two mutant cell lines after 7 days of differentiation. The heterozygous SHANK2 mutant line 6H6 showed significantly reduced amounts of the pre- and postsynaptic proteins SYP and PSD-95, whereas other proteins were not affected (Fig. 4, Supplementary Fig. 8). The compound heterozygous SHANK2 mutant line 6A4 had highly increased Nestin levels and significantly reduced levels of SYP and PSD-95, indicating impaired neuronal differentiation (Fig. 4, Supplementary Fig. 8). No differences in protein expression could be detected for β3-Tubulin, GRIN2B and AKT (Fig. 4, Supplementary Fig. 8). With β3-Tubulin immunocytofluorescence microscopy staining, we detected long dendritic protrusions starting to form networks after 7 days of differentiation in the controls (WT, 1F11) and the heterozygous SHANK2-mutant cell line 6H6 (Fig. 5a). The compound heterozygous SHANK2 cell line 6A4 lost this ability, showing less and shorter dendritic protrusions (Fig. 5a). As Nestin expression showed the most prominent change on transcriptome level in this cell line, we further investigated its expression by immunofluorescence microscopy and performed a co-staining of Nestin and β3-Tubulin during neuronal differentiation (Fig. 5b). We compared cell line 6A4 to the control 1F11 in the undifferentiated stage and after 7 and 16 days of differentiation. Both cell lines started to differentiate into neurons and Nestin-positive cells disappeared in the control cell line 1F11, whereas Nestin-positive cells could still be detected throughout differentiation in cell line 6A4. The Nestin-positive cells showed a different morphology with less cellular protrusions compared to β3-Tubulin-positive cells, even at an advanced stage of differentiation (Fig. 5b).
Immunofluorescence microscopy of SHANK2 mutant cells. (a) β3-Tubulin expression after 7 days of differentiation (DIF7) illustrating the morphology of control and SHANK2 mutant cell lines. (b) Nestin and β3-Tubulin co-staining using immunofluorescence microscopy. During 16 days of neuronal differentiation, the number of cells showing Nestin expression decreased in the control line (1F11), and nearly all cells developed neuronal morphology. In comparison, cells expressing Nestin could be detected throughout the differentiation period in the genome edited cell line (6A4). UN undifferentiated, DIF7 7 days of differentiation, DIF16 16 days of differentiation.
SHANK2 mutations affect tyrosine kinase receptor downstream signaling pathways
It has been hypothesized that insulin signaling contributes to the development of autism in genetically susceptible individuals15. As SHANK2 (as well as SHANK1 and SHANK3) directly interacts with IRSp53 (insulin receptor substrate p53), it may also play a role in insulin signaling in the brain17,37. The serine/threonine kinase AKT acts downstream of the tyrosine kinase receptor signaling pathways. Therefore, we analyzed the phosphorylation levels of AKT in relation to overall AKT levels by Western blot analysis to get an insight into a putative impairment of insulin or, more generally, into tyrosine kinase receptor downstream signaling during early neuronal differentiation. Interestingly, the cells with bi-allelic SHANK2 mutation showed an increase of phosphorylated AKT, (Fig. 6a, Supplementary Fig. 9), suggesting that a loss of SHANK2 impairs downstream signaling of tyrosine kinase receptors such as insulin receptors during early neuronal differentiation.
Analysis of the influence of SHANK2 mutations on AKT serine/threonine kinase (AKT) and amyloid precursor protein (APP) expression (7 days of differentiation) (a) and Overview of the putative influence of SHANK2 mutations on tyrosine kinase receptor downstream signaling (b). (a) Western blot analysis was carried out to identify p-AKT levels, as an indicator for tyrosine kinase receptor signaling pathway activity, and APP expression levels. P-AKT and AKT levels were normalized against GAPDH and the ratio between the normalized p-AKT/AKT expression calculated. APP expression was normalized against GAPDH. The mean for both controls (WT and 1F11) was taken for comparison with the SHANK2 mutant lines. The cell line with compound heterozygous SHANK2 mutation (6A4) showed impaired p-AKT levels and APP expression levels. Cropped western blot membrane images are shown and full-length blots are presented in Supplementary Fig. 9. n = 5 experiments, one-way ANOVA (ANOVA results: p-AKT F = 16.65, P = 0.0003; APP F = 25.83, P < 0.0001). Each mutant line was compared against the mean of both controls. ***P ≤ 0.001, correction for multiple testing with Dunnett’s test. Data were presented as box-plots. (b) Scheme illustrating the potential regulation of AKT signaling in the differentiating SH-SY5Y cells downstream of BDNF/TRKB or insulin/insulin receptor signaling. Increased p-AKT levels probably led to reduced apoptosis, increased proliferation and reduced amounts of amyloid precursor protein (APP). Further investigation is needed regarding the ERK and mTOR signaling downstream of AKT, to elucidate an effect from SHANK2 mutations. These pathways have been already linked to ASD pathology61. The part of the signaling cascade which was analyzed in this study is illustrated in green. In this simplified scheme not all different factors which influence AKT and APP expression can be considered. BDNF brain derived neurotropic factor, TRKB tyrosine receptor kinase beta, ERK extracellular-signal regulated kinase, mTOR mammalian target of rapamycin.
Bi-allelic SHANK2 mutations influence amyloid precursor protein expression
APP regulates both synapse formation and neuronal plasticity and is expressed during neuronal differentiation23,24,25. Mutations in APP have been linked to AD22,23, in which SHANK expression was also found to be altered11. SHANK2 and APP function might be linked, as they are both implicated in synaptic signal transduction, and might interact during the course of neurodegenerative disorders. Therefore, we asked if SHANK2 mutations can influence APP expression. The quantification of APP protein expression by Western blot analysis revealed a significant decrease of APP levels in the compound heterozygous SHANK2 mutant line, whereas cells with heterozygous SHANK2 mutation showed no significant difference compared to controls (Fig. 6a, Supplementary Fig. 9).
Discussion
In this study, the molecular and cellular consequences of mono-allelic and bi-allelic SHANK2 frameshift mutations were investigated in the neuroblastoma cell line SH-SY5Y. The mutations were introduced by CRISPR/Cas9 genome editing in exon 23—encoding for part of the proline-rich region—in proximity to a previously identified nonsense mutation from a patient with ASD (R841X)4. Deleterious de novo SHANK2 mutations have always been identified in a heterozygous state in patients with ASD, intellectual disability, attention deficit and hyperactivity disorder as well as language impairment, with one allele unaffected2,3,4,5. Therefore, a heterozygous SHANK2 mutant line is the appropriate model to identify impairments that contribute to the pathology in patients. Nevertheless, the compound heterozygous line is useful to explore SHANK2 function in human neurons. Our data showed that both mono- and bi-allelic mutations of SHANK2 led to reduced wildtype SHANK2 protein levels in SH-SY5Y cells. The bi-allelic mutation of SHANK2 in SH-SY5Y cells initially resulted in strongly reduced SHANK2 mRNA levels compared to the control in the undifferentiated state which is in line with reduced protein levels. After 7 days of differentiation SHANK2 mRNA levels were equal between the cell lines even though protein levels were strongly reduced and after 16 days of differentiation even an increase of SHANK2 mRNA expression was found for the compound heterozygous cell line (Supplementary Fig. 4). This might be caused by a compensatory mechanism taking place during neuronal differentiation leading to an upregulation of gene transcription because the protein is missing. The treatment of the cells during differentiation with retinoic acid and BDNF might also stimulate SHANK2 transcription.
The parallel analysis of both the compound heterozygous and the heterozygous lines revealed shared and distinct alterations in cell function. Similar consequences in both cell lines were identified concerning lower apoptosis rates, and reduced pre- and postsynaptic protein levels.
At the beginning of neuronal differentiation, cells have to switch from proliferation to differentiation38. Both SHANK2 mutant lines had lower apoptosis rates after 7 days of differentiation, and the compound heterozygous line also showed higher proliferation compared to controls. This may point to a role of SHANK2 regarding the switch from proliferation to differentiation during the early neuronal differentiation process in neuroblastoma cells. Both cell lines showed diminished pre- (SYP) and postsynaptic (PSD95) protein expression after 7 days of differentiation. SHANK2 is known to interact with PSD95 through the guanylate kinase-associated protein (GKAP) to anchor the AMPA and NMDA receptors at the postsynaptic density39. Glutamatergic markers were affected by SHANK2 mutations with reduced PSD95 and GRIA2 expression. So far, the role of SHANK2 has been mainly investigated in mature synapses30, whereas our results suggest that it may already have important functions during early neuronal differentiation, even prior to synapse formation. Interestingly, despite being a predominantly postsynaptic protein, the SHANK2 mutations also affect presynaptic protein expression, as shown by diminished SYP expression.
In the cell line with bi-allelic SHANK2 mutations expression levels of other autism-related genes and proteins (according to the SFARI Gene database6) were affected, such as SHANK1, SHANK3, GABRB3, GRIA2 and PSD95. This may point to synergistic effects and also emphasizes the importance of these genes and proteins—as well as the signaling cascades they belong to—in the pathology of autism. Cells with bi-allelic SHANK2 mutations remained in a neuronal precursor cell state, as shown by high Nestin levels and impaired neurite outgrowth. They lost their ability to extend long neurite protrusions and to form networks, which is in line with diminished levels of the pre- and postsynaptic proteins SYP and PSD95. All these observations indicate a severe impairment of neuronal differentiation. The strongest effects of Shank2 deletions were only seen in the cell line with bi-allelic SHANK2 mutation, which compares to the rat and mouse models that show the strongest phenotype only when both Shank2 alleles are deleted27,28,29. In humans bi-allelic mutations have not yet been described and may lead to embryonic death. Human individuals with heterozygous deleterious de novo SHANK2 mutations show a general developmental delay with delayed development of speech as well as learning impairments and mild to severe intellectual disability4,5. This may be a long term consequence of impaired neuronal differentiation which cannot be fully compensated. In the homozygous Shank2 knockout mice, cognitive impairments as well as a disturbed synaptic signal transmission have been observed28,29. The very early neuronal development in utero or during the first postnatal days has not been studied in these animals, therefore we do not know if an impaired neuronal differentiation may precede impaired synaptic signaling.
In a recent publication, the SHANK2-R841X mutation was investigated in human iPSC-derived cortical neurons31. The authors also showed that SHANK2-deficiency had an impact on dendritic growth, resulting in increased dendritic length and complexity, and led to a perturbation of neurodevelopmental gene set expression31. This is in line with some of our findings, even though the data between the two studies cannot be directly compared. The work on human cortical neurons (HCN2) by Luo et al.40 showed that reduced SHANK2 expression decreased neurite numbers and length in human neurons, which is comparable to our findings of reduced neurite outgrowth in the compound heterozygous SHANK2 mutant line40. They also report an impact on proliferation and apoptosis, but in their study, low SHANK2 expression inhibited proliferation and promoted apoptosis, whereas increased proliferation and decreased apoptosis was found in our study. The differences between our work and those two studies can be explained by two facts: (a) the cell models used are notably different, despite the fact that all models are human cell lines; (b) the focus of our research was on the onset of neuronal differentiation, whereas Zaslavsky et al.31 and Luo et al.40 used differentiated cortical neurons.
The severe impairment of neuronal differentiation might be partly explained by an increase of tyrosine kinase receptor signal activity. Interestingly, 6A4 cells showed elevated p-AKT levels, indicating more active tyrosine kinase receptor downstream signaling. During the process of neuronal differentiation, cells were constantly treated with retinoic acid (RA) and brain-derived neurotropic factor (BDNF). RA is used to activate the transcription of gene sets to induce differentiation and inhibit cell proliferation38. BDNF binds to tyrosine receptor kinase beta and activates several signaling cascades including the phosphatidyl-inositol-3-kinase (PI3K)—AKT pathway, which, in turn, inhibits apoptosis, regulates protein synthesis and remodels the cytoskeleton41. In parallel, other growth factors like insulin are present in the culture medium, which activate further tyrosine kinase receptors like the insulin receptor. At synapses, the insulin receptor was found co-expressed with insulin receptor substrate p53 (IRSp53), which is phosphorylated upon stimulation with insulin and acts as key factor in cytoskeleton reorganization, hence mediating neurite outgrowth42. IRSp53 is a direct interaction partner of the SHANK proteins16, linking SHANK2 to insulin signaling. As we identified increased p-AKT levels in the 6A4 cell line, which occurred together with reduced apoptosis rates and increased proliferation, we concluded that bi-allelic SHANK2 mutations might affect BDNF and/or insulin downstream signaling during early neuronal differentiation (Fig. 6b, Table 2). This is in line with other findings that describe the dysregulation of AKT signaling, which is part of the PI3K-AKT-mTor cascade downstream of tyrosine kinase receptor signaling, as a cause of ASD and other neurodevelopmental disorders43,44.
In addition, we could also show that SHANK2 mutations have an impact on APP expression, a protein extensively studied in the context of AD, although its exact biological function is not yet well understood23. The reduced APP expression in cells with bi-allelic SHANK2 mutations may also contribute to the diminished neurite outgrowth and some of the neuronal differentiation impairments. These results point to a functional link between APP and SHANK2 during neuronal development. Low APP expression occurred together with high p-AKT levels in the cell line 6A4. APP expression is known to be regulated by growth factors like insulin and BDNF which activate PI3K and AKT. AKT further phosphorylates glycogen synthase kinase 3 (GSK3) which acts as an inhibitor and results in less translation of APP protein45 (Fig. 6b).
There are functional aspects of APP which make it interesting to look at in neurodevelopmental disorders: APP regulates synapse formation, neurite outgrowth and neuronal plasticity. Furthermore, it is expressed during early neuronal differentiation at sites where SHANK2 is also expressed23,24,25,46. In addition, there is increasing evidence showing that molecular and cellular processes involved in neurodevelopment are also relevant to the pathology of neurodegenerative diseases47,48. One connection could be the pathological reactivation during neurodegeneration of genetic programs physiologically relevant during neurodevelopment, especially in Alzheimer’s disease49. There is also a genetic overlap between intellectual disability and cognitive aging48. Since SHANK2 is involved in fundamental molecular and cellular processes in neurons, it is plausible that its role might also be relevant in neurodegeneration.
The results of our study should be considered with prudence, as they were obtained in vitro in SH-SY5Y cells. As these neuroblastoma cells were tumor-derived, it is not possible to investigate all aspects of neuronal properties. The intrinsic biological variation of the neuroblastoma cell population is likely to mask low effects resulting from SHANK2 mutations, especially in the heterozygous state. Our marker gene expression analysis revealed significant expression differences, mostly in the compound heterozygous SHANK2 mutant line. The cell line with a heterozygous SHANK2 mutation may still show small changes that could not be identified due to the given limitations of our experimental setup. Despite the fact that three independent experiments were carried out, this may not allow the detection of alterations with very low effect sizes.
p-AKT levels were investigated as a general indicator of tyrosine receptor downstream signaling. Additional work is necessary to further specify which tyrosine kinase receptor signaling pathway is mostly influenced (e.g. BDNF, insulin, IGF1), which downstream signaling path of PI3K-AKT is mainly altered (e.g. mTOR, GSK3beta, ERK), and how this affects both the transcriptome and proteome.
In summary, we have shown that mono- and bi-allelic SHANK2 frameshift mutations resulted in an impairment of early neuronal differentiation in SH-SY5Y cells, characterized by changes in cell growth properties and reduced pre- and postsynaptic protein expression. We also provided the first link between SHANK2 mutations and impaired receptor tyrosine kinase downstream signaling, and revealed an impact of SHANK2 mutations on APP expression. Further investigation is warranted, preferably in human iPSC-derived neuronal cells from patients with SHANK2 mutations, to follow-up these novel insights into SHANK2-dependent pathways.
Methods
Cell culture
Human neuroblastoma cells (SH-SY5Y) were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. The cells were grown on 75 cm flasks in Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific), supplemented with 15% fetal calf serum, 1% non-essential amino acids and 1% Penicillin–streptomycin at 37 °C in a humidified environment with 5% CO2. Cells were split at 80–90% confluency and 2 × 105 cells were plated per well on a 6-well cell culture plate for RNA and protein isolation. Undifferentiated cells were harvested 3 days after seeding. Cells were differentiated according to the protocol published by Chiocchetti et al.32 and harvested after 7 or 16 days of differentiation, with medium changes 3 times per week. For neurite tracing and immunocytochemistry, 7.5 × 104 cells were seeded per well on a 24-well plate on glass coverslips coated with collagen. The cells were tested against mycoplasma contamination with the Venor GeM Classic mycoplasma detection kit for conventional PCR (Minerva Biolabs) and all used cell lines were free of mycoplasma. Genome edited cells were used for experiments with passages ranging from 15 to 20.
Genome editing
The SHANK2-gRNA was cloned with two primers: SHANK2_G2FCACCGTCGAGGGATGCCCAGAAACG and SHANK2_G2R AAACCGTTTCTGGGCATCCCTCGAC into the pSpCas9(BB)-2A-GFP vector (Addgene, 48138) using the restriction site BBsI. The plasmid was then introduced into the SH-SY5Y cells using lipofectamine (Invitrogen). Four days after transfection, GFP-positive living cells were sorted by fluorescence-activated cell sorting (FACS) as single cells into 96 well plates. The cell sorter used was a BD FACS Aria II SORP system. After 1–3 months, colonies could be split and analyzed for mutations. Medium changes were performed weekly.
Quantitative real time PCR (qPCR)
Total RNA from SH-SY5Y cells was extracted with the RNAqueous—Micro Kit (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). qPCR was conducted using the SYBR Green Lo-Rox Fast Mix (Bioline) and the ABI 7500 Fast Real-Time PCR system (Applied Biosystems). Each sample was analyzed in triplicates. Relative mRNA levels were assessed using the relative standard curve method by normalization to the mean of following reference RNAs: glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, heat shock protein family D (HSP60) member1 (HSPD1) mRNA and hypoxanthine phosphoribosyltransferase 1 mRNA (HPRT1)50. The mean value and the standard error of the mean (SEM) of the relative expression values are presented. The sequences for the oligonucleotides used are given in Supplementary Table 2.
Nonsense mediated mRNA decay
2 × 105 cells were seeded per well on a 6-well plate and cultured for 24 h. Then, culturing medium was changed to either mock treatment (2 ml medium + 50 µl water) or cycloheximide (CHX) treatment (100 µg/ml, CHX was dissolved in water) and cultured for 6 h. Then, cells were harvested, RNA was isolated and SHANK2 expression quantified with the primers SHANK2_F and SHANK2_R (details in Supplementary Table 2). Values of three independent experiments were presented as box-plots.
Sequencing
DNA was isolated from SH-SY5Y cells with the Quick-DNA Miniprep kit (ZYMO), the region flanking the SHANK2 mutation site was amplified with primers SH2_RXCheck_F agctgcagcgaataggaaag and SH2_RXCheck_R tgggcttcaagatgacagaa with Hotstar Taq (Qiagen) and sent for Sanger sequencing (GENEWIZ, Leipzig, Germany).
Immunofluorescence microscopy
Immunocytofluorescence analysis was performed on fixed SH-SY5Y cells, treated with 4% paraformaldehyde solution in PBS for 15 min at room temperature, using the primary antibodies anti-β3-Tubulin51 (Abcam, ab18207, knockout validated, 1:500 dilution), anti-Nestin (Abcam, ab22035, 1:100 dilution) or anti-Ki6752 (Abcam, ab16667, knockout validated, 1:500 dilution), and as secondary antibodies Alexa Fluor 488 goat anti-rabbit, Alexa Fluor 488 donkey anti-rabbit, Alexa Fluor 546 rabbit anti-mouse or Alexa Fluor 546 goat anti-mouse (Thermo Fisher Scientific, 1:1000 dilution). Nuclei were stained with Hoechst dye (Hoechst 33,342). The TUNEL assay was carried out with the in situ Cell Death Detection kit, Fluorescein (Roche) according to the manufacturer’s instructions.
For the analysis of neurite length, cells were fixed after 4 days of differentiation. Images were taken in the channel corresponding to the β3-Tubulin signal and neurites were traced using the “Simple neurite tracer” tool of the FiJi software36 to determine their length. Three independent experiments were carried out and cells from six images were analyzed for each cell line per experiment.
Ki67 and TUNEL signals were quantified by using the FiJi36 software and by normalizing the number of signals to the total number of nuclei (as determined by Hoechst 33342 staining). Three independent experiments were carried out and cells of six images were analyzed for each cell line per experiment.
For the follow-up of Nestin expression levels, three independent experiments were performed, and 6 representative images were selected per experiment. All coverslips were mounted using Aqua-Poly/Mount medium (Polysciences).
Protein analysis
Protein extraction from SH-SY5Y cells was performed at 4 °C using RIPA buffer supplemented with SIGMAFAST protease inhibitor (S8820; Sigma). Protein concentrations were determined with the BCA protein assay kit (Pierce, Thermo Scientific). Western blot analysis was executed using the Odyssey Infrared Imaging System (LI-COR Biosciences). Twenty-five micrograms of proteins were separated on Novex WedgeWell 4–12% Tris Glycine Gels (Thermo Fisher Scientific) and transferred to PVDF membranes (Millipore). In a pilot experiment, protein concentrations from 15 to 40 µg were loaded which revealed that the amount of 25 µg protein per sample is in a linear quantification range for all analyzed proteins (Supplementary Figs. 10 and 11). PVDF membranes (Millipore) were probed with rabbit polyclonal β3-Tubulin53 (1:20,000; Abcam, ab18207, knockout validated), sheep anti-SHANK231 (1:2,500; R&D Systems, AF7035), rabbit anti-GRIN2B54 (1:1,000; Abcam, ab65783), mouse anti-PSD9555,56 (1:500; NeuroMab, 75–028, knockout validated), rabbit anti-Synaptophysin57 (1:7,500; Abcam, ab32127, knockout validated), mouse anti-GAPDH58 (1:10,000; Novus Biologicals, NB300-328), mouse anti-Nestin59 (1:2,500; Abcam, ab22035), rabbit anti-AKT45 (1:1,000; Cell Signaling Technology, 9272), mouse anti p-AKT(Ser473)45,60 (1:1,000; Cell Signaling Technology, 4051) and rabbit anti-APP45 (1:500, Abcam, ab126732, knockout validated).
The secondary antibodies used were IRDye 800CW donkey anti-mouse, IRDye 680LT donkey anti-mouse, IRDye 680RD donkey anti-rabbit, IRDye 800CW donkey anti-rabbit or IRDye 800CW donkey anti-goat (1:15,000 dilution; LI-COR Biosciences). Immuno-positive signals were quantified using the Image Studio Lite 3.1 software (LI-COR Biosciences). The Page Ruler Prestained Protein ladder (10–180 kDa) and the Spectra Multicolor High Range Protein ladder (40–300 kDa; both Thermo Fisher Scientific) were used as protein size markers.
Protein expression was normalized to the amount of GAPDH and the values obtained for the reference line 1F11 were set to 100. P-AKT and AKT levels were first normalized against GAPDH and then the ratio of the normalized data P-AKT/AKT was calculated. The normalized values were presented in box-plots.
Equipment and settings
Westernblot images in Figs. 1, 4 and 6 were acquired with the Image Studio Software 4.0 (LICOR) with the scanning intensity of 5.0 for channel 700 and of 4.5 for channel 800.
Immunofluorescence images in Fig. 2 were taken with the Leica DMI4000B microscope with the camera Leica-DFC3000G-0001062113 and the objective N PLAN L 20×/0.35 DRY using the Leica Application Suite X software in the red, green and blue channel. The acquisition was performed with the time and space resolution XY = 0.959 µm and Z = 4.490 µm, 12 bit images were taken with a resolution of 1296 × 966 pixels. All pictures were taken with the same settings. In the illustrated TUNEL and Ki67 pictures brightness was increased by 30% for all conditions in the Power Point software. Images in Fig. 3 were taken with the same microscope and the same settings only for the green channel, brightness was increased by 20% for all conditions in Power Point software.
Images in Fig. 5 were taken with the same microscope and camera and the objective HC PL APO 40×/0.85 DRY. The acquisition was performed with the time and space resolution XY = 0.395 µm and Z = 0.761 µm, 12 bit images were taken with a resolution of 1296 × 966 pixel. All pictures were taken with the same settings. Colour saturation was increased to 200% for all conditions in Power Point software.
Statistical analysis
Prism 8 (GraphPad Software) software was used for data analysis. The Shapiro Wilk test was performed to test for normal distribution when sample sizes were ≥ 5. In the TUNEL assay, Ki67 quantification, analysis of neurite length and Western blot experiments, a one-way ANOVA test was used for the comparison of each SHANK2 mutant line (6A4, 6H6) against the reference 1F11. In addition, the WT line was compared to 1F11. A P-value of ≤ 0.05 was considered nominally significant. The Dunnett’s test was used to correct for multiple testing (n = 3 tests). A two-tailed unpaired Student´s T-Test was performed for the pairwise comparison of protein expression in undifferentiated versus the 7 days differentiated WT cell line.
Two-way ANOVA was performed for the comparison of RNA expression levels between the reference 1F11 and the two mutant lines (6A4, 6H6), with gene and cell line as the two factors; the Dunnett’s test was used to correct for multiple testing for each time point.
Data availability
The datasets supporting the conclusions of this article are included within the article and its additional files.
References
Leblond, C. S. et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: A gradient of severity in cognitive impairments. PLoS Genet. 10, e1004580. https://doi.org/10.1371/journal.pgen.1004580 (2014).
Bowling, K. M. et al. Genomic diagnosis for children with intellectual disability and/or developmental delay. Genome Med. 9, 43. https://doi.org/10.1186/s13073-017-0433-1 (2017).
Yuen, R. K. et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602–611. https://doi.org/10.1038/nn.4524 (2017).
Berkel, S. et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 42, 489–491. https://doi.org/10.1038/ng.589 (2010).
Leblond, C. S. et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 8, e1002521. https://doi.org/10.1371/journal.pgen.1002521 (2012).
Abrahams, B. S. et al. SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36. https://doi.org/10.1186/2040-2392-4-36 (2013).
Peykov, S. et al. Identification and functional characterization of rare SHANK2 variants in schizophrenia. Mol. Psychiatry 20, 1489–1498. https://doi.org/10.1038/mp.2014.172 (2015).
Noor, A. et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Society Psychiatr. Genet. 165B, 303–313. https://doi.org/10.1002/ajmg.b.32232 (2014).
Homann, O. R. et al. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol. Psychiatry 21, 1690–1695. https://doi.org/10.1038/mp.2016.24 (2016).
Pappas, A. L. et al. Deficiency of Shank2 causes mania-like behavior that responds to mood stabilizers. JCI Insight https://doi.org/10.1172/jci.insight.92052 (2017).
Gong, Y., Lippa, C. F., Zhu, J., Lin, Q. & Rosso, A. L. Disruption of glutamate receptors at Shank-postsynaptic platform in Alzheimer’s disease. Brain Res. 1292, 191–198. https://doi.org/10.1016/j.brainres.2009.07.056 (2009).
Sheng, M. & Sala, C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24, 1–29. https://doi.org/10.1146/annurev.neuro.24.1.1 (2001).
Boeckers, T. M., Bockmann, J., Kreutz, M. R. & Gundelfinger, E. D. ProSAP/Shank proteins—A family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J. Neurochem. 81, 903–910. https://doi.org/10.1046/j.1471-4159.2002.00931.x (2002).
Monteiro, P. & Feng, G. SHANK proteins: Roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci. 18, 147–157. https://doi.org/10.1038/nrn.2016.183 (2017).
Stern, M. Insulin signaling and autism. Front. Endocrinol. (Lausanne) 2, 54. https://doi.org/10.3389/fendo.2011.00054 (2011).
Bockmann, J., Kreutz, M. R., Gundelfinger, E. D. & Bockers, T. M. ProSAP/Shank postsynaptic density proteins interact with insulin receptor tyrosine kinase substrate IRSp53. J. Neurochem. 83, 1013–1017. https://doi.org/10.1046/j.1471-4159.2002.01204.x (2002).
Kim, R. et al. Cell-type-specific Shank2 deletion in mice leads to differential synaptic and behavioral phenotypes. J. Neurosci. 38, 4076–4092. https://doi.org/10.1523/JNEUROSCI.2684-17.2018 (2018).
Bozdagi, O., Tavassoli, T. & Buxbaum, J. D. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol. Autism 4, 9. https://doi.org/10.1186/2040-2392-4-9 (2013).
Shcheglovitov, A. et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature 503, 267–271. https://doi.org/10.1038/nature12618 (2013).
Chen, S. T. et al. Insulin-like growth factor 1 partially rescues early developmental defects caused by SHANK2 knockdown in human neurons. Neural Regen. Res. 15, 2335–2343. https://doi.org/10.4103/1673-5374.285002 (2020).
Guilmatre, A., Huguet, G., Delorme, R. & Bourgeron, T. The emerging role of SHANK genes in neuropsychiatric disorders. Develop. Neurobiol. 74, 113–122. https://doi.org/10.1002/dneu.22128 (2014).
O’Brien, R. J. & Wong, P. C. Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 34, 185–204. https://doi.org/10.1146/annurev-neuro-061010-113613 (2011).
van der Kant, R. & Goldstein, L. S. Cellular functions of the amyloid precursor protein from development to dementia. Develop. Cell 32, 502–515. https://doi.org/10.1016/j.devcel.2015.01.022 (2015).
Priller, C. et al. Synapse formation and function is modulated by the amyloid precursor protein. J. Neurosci. 26, 7212–7221. https://doi.org/10.1523/JNEUROSCI.1450-06.2006 (2006).
Turner, P. R., O’Connor, K., Tate, W. P. & Abraham, W. C. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog. Neurobiol. 70, 1–32. https://doi.org/10.1016/s0301-0082(03)00089-3 (2003).
Berkel, S. et al. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum. Mol. Genet. 21, 344–357. https://doi.org/10.1093/hmg/ddr470 (2012).
Modi, M. E. et al. Hyperactivity and hypermotivation associated with increased striatal mGluR1 signaling in a Shank2 rat model of autism. Front. Mol. Neurosci. 11, 107. https://doi.org/10.3389/fnmol.2018.00107 (2018).
Schmeisser, M. J. et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 486, 256–260. https://doi.org/10.1038/nature11015 (2012).
Won, H. et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 486, 261–265. https://doi.org/10.1038/nature11208 (2012).
Eltokhi, A., Rappold, G. & Sprengel, R. Distinct phenotypes of Shank2 mouse models reflect neuropsychiatric spectrum disorders of human patients with SHANK2 variants. Front. Mol. Neurosci. 11, 240. https://doi.org/10.3389/fnmol.2018.00240 (2018).
Zaslavsky, K. et al. SHANK2 mutations associated with autism spectrum disorder cause hyperconnectivity of human neurons. Nat. Neurosci. 22, 556–564. https://doi.org/10.1038/s41593-019-0365-8 (2019).
Chiocchetti, A. G. et al. Transcriptomic signatures of neuronal differentiation and their association with risk genes for autism spectrum and related neuropsychiatric disorders. Transl. Psychiatry 6, e864. https://doi.org/10.1038/tp.2016.119 (2016).
Berkel, S. et al. Sex hormones regulate SHANK expression. Front. Mol. Neurosci. 11, 337. https://doi.org/10.3389/fnmol.2018.00337 (2018).
Schumann, C. M. et al. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J. Neurosci. 30, 4419–4427. https://doi.org/10.1523/JNEUROSCI.5714-09.2010 (2010).
Stoner, R. et al. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 370, 1209–1219. https://doi.org/10.1056/NEJMoa1307491 (2014).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. https://doi.org/10.1038/nmeth.2019 (2012).
Chow, F., Gong, Y. & Lippa, C. F. The potential role of insulin on the shank-postsynaptic platform in neurodegenerative diseases involving cognition. Am. J. Alzheimers Dis. Other Demen. 29, 303–310. https://doi.org/10.1177/1533317513518645 (2014).
Janesick, A., Wu, S. C. & Blumberg, B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. CMLS 72, 1559–1576. https://doi.org/10.1007/s00018-014-1815-9 (2015).
Boeckers, T. M. et al. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem. Biophys. Res. Commun. 264, 247–252. https://doi.org/10.1006/bbrc.1999.1489 (1999).
Luo, T. et al. Effect of the autism-associated lncRNA Shank2-AS on architecture and growth of neurons. J. Cell. Biochem. https://doi.org/10.1002/jcb.27471 (2018).
Kowianski, P. et al. BDNF: A Key factor with multipotent impact on brain signaling and synaptic plasticity. Cell. Mol. Neurobiol. 38, 579–593. https://doi.org/10.1007/s10571-017-0510-4 (2018).
Blazquez, E., Velazquez, E., Hurtado-Carneiro, V. & Ruiz-Albusac, J. M. Insulin in the brain: Its pathophysiological implications for States related with central insulin resistance, type 2 diabetes and Alzheimer’s disease. Front. Endocrinol. (Lausanne) 5, 161. https://doi.org/10.3389/fendo.2014.00161 (2014).
Wang, L. et al. Brain development and Akt signaling: The crossroads of signaling pathway and neurodevelopmental diseases. J. Mol. Neurosci. 61, 379–384. https://doi.org/10.1007/s12031-016-0872-y (2017).
Winden, K. D., Ebrahimi-Fakhari, D. & Sahin, M. Abnormal mTOR activation in autism. Annu. Rev. Neurosci. 41, 1–23. https://doi.org/10.1146/annurev-neuro-080317-061747 (2018).
Chen, L. et al. Methamphetamine exposure upregulates the amyloid precursor protein and hyperphosphorylated tau expression: The roles of insulin signaling in SH-SY5Y cell line. J. Toxicol. Sci. 44, 493–503. https://doi.org/10.2131/jts.44.493 (2019).
Dawkins, E. & Small, D. H. Insights into the physiological function of the beta-amyloid precursor protein: Beyond Alzheimer’s disease. J. Neurochem. 129, 756–769. https://doi.org/10.1111/jnc.12675 (2014).
Parikshak, N. N., Gandal, M. J. & Geschwind, D. H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458. https://doi.org/10.1038/nrg3934 (2015).
Starr, J. M. Ageing and epigenetics: Linking neurodevelopmental and neurodegenerative disorders. Dev. Med. Child Neurol. 61, 1134–1138. https://doi.org/10.1111/dmcn.14210 (2019).
Bothwell, M. & Giniger, E. Alzheimer’s disease: Neurodevelopment converges with neurodegeneration. Cell 102, 271–273. https://doi.org/10.1016/s0092-8674(00)00032-5 (2000).
Cikos, S., Bukovska, A. & Koppel, J. Relative quantification of mRNA: Comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 8, 113. https://doi.org/10.1186/1471-2199-8-113 (2007).
Saric, N. et al. The AHR pathway represses TGFbeta-SMAD3 signalling and has a potent tumour suppressive role in SHH medulloblastoma. Sci. Rep. 10, 148. https://doi.org/10.1038/s41598-019-56876-z (2020).
Taylor-Whiteley, T. R., Le Maitre, C. L., Duce, J. A., Dalton, C. F. & Smith, D. P. Recapitulating Parkinson’s disease pathology in a three-dimensional human neural cell culture model. Dis. Model Mech. https://doi.org/10.1242/dmm.038042 (2019).
Spilsbury, A., Miwa, S., Attems, J. & Saretzki, G. The role of telomerase protein TERT in Alzheimer’s disease and in tau-related pathology in vitro. J. Neurosci. 35, 1659–1674. https://doi.org/10.1523/JNEUROSCI.2925-14.2015 (2015).
Colas, J. et al. Neuroprotection against amyloid-beta-induced DNA double-strand breaks is mediated by multiple retinoic acid-dependent pathways. Neural Plast. 2020, 9369815. https://doi.org/10.1155/2020/9369815 (2020).
Gutierrez, D. A. et al. c-Abl deficiency provides synaptic resiliency against abeta-oligomers. Front. Cell Neurosci. 13, 526. https://doi.org/10.3389/fncel.2019.00526 (2019).
Park, H. et al. Splice-dependent trans-synaptic PTPdelta-IL1RAPL1 interaction regulates synapse formation and non-REM sleep. EMBO J. 39, e104150. https://doi.org/10.15252/embj.2019104150 (2020).
Linker, K. E. et al. Microglial activation increases cocaine self-administration following adolescent nicotine exposure. Nat. Commun. 11, 306. https://doi.org/10.1038/s41467-019-14173-3 (2020).
Huguier, V. et al. Oncostatin M exerts a protective effect against excessive scarring by counteracting the inductive effect of TGFbeta1 on fibrosis markers. Sci. Rep. 9, 2113. https://doi.org/10.1038/s41598-019-38572-0 (2019).
Cai, S. et al. Directed differentiation of human bone marrow stromal cells to fate-committed schwann cells. Stem Cell Rep. 9, 1097–1108. https://doi.org/10.1016/j.stemcr.2017.08.004 (2017).
Zhang, Z. et al. TLR4 counteracts BVRA signaling in human leukocytes via differential regulation of AMPK, mTORC1 and mTORC2. Sci. Rep. 9, 7020. https://doi.org/10.1038/s41598-019-43347-8 (2019).
Rosina, E. et al. Disruption of mTOR and MAPK pathways correlates with severity in idiopathic autism. Transl. Psychiatry 9, 50. https://doi.org/10.1038/s41398-018-0335-z (2019).
Acknowledgements
We would like to thank Zeynep Aydin, Diana Porras Gonzalez, Ahmed Eltokhi and Andrew Lindsey for experimental help, and Beatrix Startt for editing support. This study was supported by the Medical Faculty of the Ruprecht-Karls-University Heidelberg. SB was supported by a Frontier grant (ZUK49/Ü5.2.183) and the Olympia Morata program of Heidelberg University.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
C.U. carried out the cell culture work, isolation of DNA, RNA and protein, immunofluorescence microscopy, Western blot analysis, qPCR, sequencing and analyzed the data. She prepared figures and contributed to the writing of the manuscript. F.-B.C. contributed to the immunocytofluorescence experiments, data analysis and writing of the manuscript. M.v.H. carried out Western blot analysis, analyzed sequences and prepared figures. V.E. performed the fluorescence activated cell sorting. G.R. contributed to the data analysis and writing of the manuscript and supported the study. S.B. designed the study, contributed to the experiments, analyzed and interpreted the data and wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Unsicker, C., Cristian, FB., von Hahn, M. et al. SHANK2 mutations impair apoptosis, proliferation and neurite outgrowth during early neuronal differentiation in SH-SY5Y cells. Sci Rep 11, 2128 (2021). https://doi.org/10.1038/s41598-021-81241-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-81241-4
This article is cited by
-
Lactate Protects Microglia and Neurons from Oxygen–Glucose Deprivation/Reoxygenation
Neurochemical Research (2024)
-
Taurine Alleviates Chronic Social Defeat Stress-Induced Depression by Protecting Cortical Neurons from Dendritic Spine Loss
Cellular and Molecular Neurobiology (2023)
-
Quantitative proteomics and in-cell cross-linking reveal cellular reorganisation during early neuronal differentiation of SH-SY5Y cells
Communications Biology (2022)
-
A perspective on molecular signalling dysfunction, its clinical relevance and therapeutics in autism spectrum disorder
Experimental Brain Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.