Abstract
The variant virulent porcine epidemic diarrhea virus (PEDV) strain (YN15) can cause severe porcine epidemic diarrhea (PED); however, the attenuated vaccine-like PEDV strain (YN144) can induce immunity in piglets. To investigate the differences in pathogenesis and epigenetic mechanisms between the two strains, differential expression and correlation analyses of the microRNA (miRNA) and mRNA in swine testicular (ST) cells infected with YN15, YN144, and mock were performed on three comparison groups (YN15 vs Control, YN144 vs Control, and YN15 vs YN144). The mRNA and miRNA expression profiles were obtained using next-generation sequencing (NGS), and the differentially expressed (DE) (p-value < 0.05) mRNA and miRNA were obtained using DESeq R package. mRNAs targeted by DE miRNAs were predicted using the miRanda algortithm. 8039, 8631 and 3310 DE mRNAs, and 36, 36, and 22 DE miRNAs were identified in the three comparison groups, respectively. 14,140, 15,367 and 3771 DE miRNA–mRNA (targeted by DE miRNAs) interaction pairs with negatively correlated expression patterns were identified, and interaction networks were constructed using Cytoscape. Six DE miRNAs and six DE mRNAs were randomly selected to verify the sequencing data by real-time relative quantitative reverse transcription polymerase chain reaction (qRT-PCR). Based on bioinformatics analysis, we discovered the differences were mostly involved in host immune responses and viral pathogenicity, including NF-κB signaling pathway and bacterial invasion of epithelial cells, etc. This is the first comprehensive comparison of DE miRNA–mRNA pairs in YN15 and YN144 infection in vitro, which could provide novel strategies for the prevention and control of PED.
Similar content being viewed by others
Introduction
As an acute enteric disease of piglets, porcine epidemic diarrhea (PED) is typically characterized by watery diarrhea accompanied by vomiting, dehydration and high mortality1. PED virus (PEDV), a member of the Coronaviridae family, is the etiological agent of PED and a single-stranded positive-sense RNA virus whose genome is approximately 28 kb in size. The genome includes at least seven open reading frames (ORFs) of which ORF1a and ORF1b encode nonstructural proteins. The other five ORFs encode four structural proteins and one accessory protein, namely spike (S) protein, ORF3, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein in sequence1.
Since the late 2010s, a severe outbreak of PED has affected Asian, Europe and the Americas, and has caused great financial losses to the swine industry throughout the world2,3,4. The variant PEDV YN1 strain was isolated, passaged to 15 generations to obtain the variant virulent strain. This strain was passaged further for a total of 144 passages to obtain the attenuated, YN1445. In animal experiments, rather than inducing diarrhea in piglets, the YN144 strain induced innate immunity and neutralizing antibody responses, and provided protection to piglets challenged with a variant virulent strain5. In order to better understand the pathogenic mechanism of the variant PEDVs, a proteomics study was performed in vivo following the variant and attenuated strain infection, whose differentially expressed proteins are associated with immune function and stress responses6.
Accumulating studies have demonstrated that microRNA (miRNA) can regulate the signaling pathways of host cells to defend against viral infections7. miRNA is a single-stranded, noncoding RNA of approximately 18–24 nucleotide length and is derived from longer preprimary transcripts8. The target recognition sequence of a mature miRNA, called the seed region, is highly conserved and is located within positions 2–8 of the miRNA. miRNA can regulate the expression of target mRNAs by binding to the 3′ or 5′ untranslated region of target mRNA using its seed region and ultimately inducing mRNA degradation, thereby playing a crucial role in controlling post-transcriptional gene expression in almost all known physiological and pathophysiological processes9. miRNA has two distinct anti-viral mechanisms. First, miRNA directly targets viral mRNA10. Second, miRNA can activate the host immune responses to control viral replication11. Next-generation sequencing (NGS) has become a powerful tool to analyze the expression profiles of miRNA and mRNA12.
In this study, the differential expression and correlation analysis of the differentially expressed (DE, p-value < 0.05) miRNA and mRNA in swine testicular (ST) cells infected with the variant virulent PEDV strain (YN15) and the attenuated vaccine-like PEDV strain (YN144) were investigated to understand the roles of the miRNA–mRNA network in PEDV pathogenesis using NGS technology. These findings could lead to novel ideas for the prevention and control of PED.
Results
Replication kinetics of PEDV variant strains YN15 and YN144 in ST cells in vitro
Viral titers in ST cells were monitored at different time points after YN15 or YN144 infection (Fig. 1). The growth curves of the two PEDV strains, YN15 and YN144, in ST cells show that both strains peak at 48 h post-infection (pi), then gradually decline (Fig. 1a). In order to make the process of replication kinetics and identification of the intracellular PEDV (YN15 and YN144 strains) more visual, indirect immunofluorescence assay (IIFA) was used to track the intracellular PEDV strains. The intracellular PEDV replication over the first three 12 h interval time points was validated, and there was a high virus yield at 24 h pi (Fig. 1b). RNA samples were prepared using the cell pellets collected at 24 h pi for further small RNA and mRNA sequencing.
The growth curves of the two PEDV strains (YN15 and YN144) in ST cells (0.001 multiplicity of infection (MOI)). (a) The growth curves of the two PEDV strains (YN15 and YN144) in ST cells. (b) Confirmation of YN15 and YN144 replication in ST cells at 12, 24, and 36 h pi by IIFA. Mock-infected ST cells at 36 h pi were set as control. Scale bar = 100 μm.
Global mRNA expression patterns in ST cells after YN15 and YN144 infection
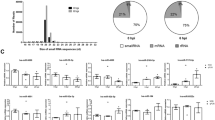
To investigate mRNA expression profiles during PEDV (YN15 or YN144) infection, gene transcripts were analyzed using RNA-seq, and the same total RNAs were used to construct nine sample sequence libraries. The quality assessment of the sequencing data is summarized in Supplementary Table S1. Among the clean reads, most were mapped to the Sus scrofa genome, and the sequence alignment results are shown in Supplementary Table S2. Genes were defined as DE when the p-value for the difference in expression was < 0.05 (padj < 0.05). Compared with the control group (uninfected ST cells), 8039 (3959 up-regulated and 4080 down-regulated) DE mRNAs were identified in the YN15 infection group and 8631 (4355 up-regulated and 4276 down-regulated) DE mRNAs were identified in the YN144 infection group. When the DE mRNAs in YN15 group were compared to those of YN144 group, 3310 (1494 up-regulated and 1816 down-regulated) DE mRNAs were commonly DE in both (Fig. 2a and Supplementary Table S3).
mRNAs expression profiles in PEDV-infected ST cells. (a) The bar graph shows the number of DE mRNAs in each one of the three comparison groups, red represents the amount of up-regulated DE mRNAs, green represents the count of down-regulated DE mRNAs (p-value < 0.05). (b) GO analysis of the DE mRNAs in PEDV infection ST. The three panels show the GO enrichment analysis of each of the top 30 significantly most DE mRNAs from the three comparison groups (YN15 vs Control, YN144 vs Control, and YN15 vs YN144), respectively. (c) KEGG analysis of the DE mRNAs in ST cells infected with PEDV. The three panels show the KEGG enrichment analysis of each of the top 20 significantly most DE mRNAs between YN15, YN144 and control groups, respectively. (d) Results of relative quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis (alongside RNA-seq analysis) of ATF3, CRAMP, NKTR, RND1, SMAGP and TNFSF10 in YN15- and YN144-infected ST cells, respectively. The relative expression of each mRNA was calculated using the 2-ΔΔCt method. The data are presented as mean ± SEM from three independent experiments.
Gene Ontology (GO)-based analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) mapping were performed to explore the biological functions of all of the DE mRNAs. The most significantly enriched GO terms for target mRNAs in the three comparison groups are shown in Fig. 2b. All of the KEGG enrichment pathways are shown in Supplementary Table S4; among them each of the top 20 significantly enriched KEGG pathways is shown in Fig. 2c. Although the DE mRNAs differ depending on the specific experimental group comparison, most of them were involved in biological process (bp), cellular component (cc) and molecular function (mf). There were some commonly enriched GO terms, including protein binding, metabolic process, organic substance metabolic process, intracellular part, and organelle. In addition to the commonly enriched GO terms, the uniquely DE mRNAs in the YN15 vs Control comparison group, are involved in extracellular exosome, membrane-bounded vesicle, extracellular organelle, and vesicle. The uniquely DE mRNAs in the YN144 vs Control comparison group are involved in macromolecule metabolic process, cellular component organization, nucleoplasm, and binding. In the YN15 vs YN144 comparison group, the top 30 enriched GO terms for the DE mRNAs are involved in immune system process, regulation of cellular process, regulation of biological process, programmed cell death, apoptotic processes, and inflammatory response. According to the KEGG mapping of the DE mRNAs, most of the top 20 significant signaling pathways were different across the three comparison groups, except for pathway in cancer, tumor necrosis factor (TNF) signaling pathway and toxoplasmosis. The DE mRNAs are mostly involved in Fc gamma R-mediated phagocytosis, fatty acid metabolism, apoptosis, AMPK signaling pathway, and bacterial invasion of epithelial cells in the YN15 vs Control comparison group. While, the DE mRNAs in the YN144 vs Control comparison group are involved in viral carcinogenesis, RIG-I-like receptor signaling pathway, NOD-like receptor signaling pathway, chronic myeloid leukemia, and endocytosis. In the YN15 vs YN144 comparison group, the DE mRNAs are mostly involved in Toll-like signaling pathway, apoptosis, extracellular matrix protein (ECM)-receptor interaction, and the nuclear factor kappa-B (NF-κB) signaling pathway.
To confirm the results obtained from RNA-seq, six randomly selected mRNAs (activating transcription factor 3 (ATF3), cramped chromatin regulator homolog (CRAMP), natural killer cell triggering receptor (NKTR), Rho family GTPase 1 (RND1), small cell adhesion glycoprotein (SMAGP) and tumor necrosis factor superfamily member 10 (TNFSF10)) were measured using real-time relative quantitative reverse transcription polymerase chain reaction (qRT-PCR) and their changes in expression showed trends consistent with the RNA-seq results (Fig. 2d), indicating that the RNA-seq data are reflective of actual changes at the genetic level.
Global miRNA expression patterns in ST cells after YN15 and YN144 infection
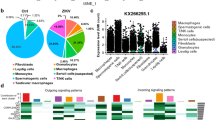
Raw reads from the libraries derived from YN15, YN144 and control groups were generated. Clean reads were obtained after filtration. The clean reads successfully mapped to the reference genome sequence of Sus scrofa, as shown in Supplementary Table S5. The length of the majority of clean reads obtained were 22–24 nucleotides (nt), and the size of 23 nt was the most common (Fig. 3a). The percentage of nucleic acid represented by miRNAs in YN15, YN144, and control groups is shown in Fig. 3b. The numbers of mapped known and novel miRNAs are listed in Supplementary Table S6. After BLASTN analysis by miRBase 20.0 with Sus scrofa miRNAs, 275 known and 279 novel mature miRNAs were obtained (Supplementary Table S7).
miRNA expression profiles in PEDV-infected ST cells. (a) Length of distribution of small RNA-seq reads. (b) Pie charts of small RNA-seq results showing the percentage of small RNA components in PEDV YN15-, YN144-, and mock-infected (Control) ST cells. (c) Volcano plots of up-regulated and down-regulated differentially expressed microRNAs (DE miRNAs) in three comparison groups (YN15 vs Control, YN144 vs Control, and YN15 vs YN144) (p-value < 0.05). (d) Venn diagram of the number of the commonly and uniquely DE miRNAs between the two comparison groups (YN15 vs Control and YN144 vs Control). Each number in a circle represents the number of the DE miRNAs found in each comparison group. The number in the overlapping area represents miRNAs commonly DE between the two comparison groups. The number in each non-overlapping area indicates the unique DE miRNAs in each comparison group. (p-value < 0.05). (e) Results of real-time relative qRT-PCR analysis (alongside RNA-seq analysis) of ssc-miR-9–1, ssc-miR-27a, ssc-miR-32, ssc-miR-192, ssc-miR-671-5p and ssc-miR-1306-5p fold-change in the two comparison groups (YN15 vs Control and YN144 vs Control). The relative expression of each miRNA was calculated using the 2-ΔΔCt method. The data are presented as mean ± SEM from three independent experiments.
Identification of DE miRNAs was performed based on p-value < 0.05. Based on the volcano plots, three DE miRNA clusters emerged (Fig. 3c). Among these DE miRNAs, YN15 infection was found to increase the expression of 15 miRNAs and decrease the expression of 21 miRNAs in ST cells, YN144 infection up-regulated 15 miRNAs and down-regulated 21 miRNAs, and 22 miRNAs, 9 up- and 13 down-regulated, were identified when comparing ST cells infected with YN15 and ST cells infected with YN144 (Fig. 3c and Supplementary Table S8). Among all of DE miRNAs, seven novel miRNAs were discovered (Supplementary Table S8).As shown in the Venn diagram (Fig. 3d), 25 commonly and 22 uniquely DE miRNAs were identified between the YN15 vs Control comparison group and the YN144 vs Control comparison group. All 25 commonly DE miRNAs exhibited the same trend in expression, i.e., 11 up-regulated and 14 down-regulated miRNAs (Supplementary Table S9).
To identify the data obtained from small RNA-seq, six randomly selected miRNAs (ssc-miR-9–1, ssc-miR-27a, ssc-miR-32, ssc-miR-192, ssc-miR-671-5p and ssc-miR-1306-5p) were evaluated by real-time relative qRT-PCR (Fig. 3e). The comparable results indicate that the data obtained by RNA-seq were valid.
Prediction and functional characterization of target mRNAs of DE miRNAs
The target mRNAs of DE miRNA identified in the three comparison groups (YN15 vs Control, YN144 vs Control, and YN15 vs YN144) were predicted using the miRanda algorithm (Table. 1). GO and KEGG analyses were performed to characterize the functions of the predicted target mRNAs. The most significantly enriched GO terms of target mRNAs between the three comparison groups are involved in the biological processes of bp, cc and mf (Fig. 4a). All of the enriched pathways identified through KEGG mapping are shown in Supplementary Table S10; among them, the top 20 significantly enriched pathways represented by predicted target mRNAs from each comparison group were identified (Fig. 4b). In the YN15 vs Control comparison group, the target mRNAs were associated with immune- and bowel disease-related signaling pathways, including T cell receptor signaling pathway, Toll-like receptor signaling pathway, and inflammatory bowel disease (IBD), etc. In the YN144 vs Control comparison group, the enriched genes were involved in immune-related signaling pathways, including T cell receptor signaling pathway, intestinal immune network for IgA production, human T-cell leukemia virus type 1 (HTLV-1) infection, Fc gamma R-mediated phagocytosis, and B cell receptor signaling pathway. When YN15 and YN144 infected cells were compared, the target mRNAs of DE miRNAs were almost all connected to immune- and bowel disease-related signaling pathways, either IBD, Rap1 signaling pathway, HTLV-1 infection, Fc gamma R-mediated phagocytosis, T cell receptor signaling pathway, or the NF-κB signaling pathway.
DE miRNAs and their predicted target mRNAs in PEDV-infected ST cells. (a) GO analysis of the predicted target genes of the DE miRNAs from the three comparison groups. (b) KEGG analysis of the predicted target genes of the DE miRNAs from the three comparison groups. Each panels indicates the top 20 significantly enriched pathways identifed by KEGG analysis of each of the three comparison groups (Left, YN15 vs Control; Middle, YN144 vs Control; Right, YN15 vs YN144).
Joint functional analysis of DE miRNAs and DE target mRNAs
Using the strategy illustrated in Fig. 5a, a large number of DE miRNA–mRNA pairs were identified (p-value < 0.05). Our work indicated that one mRNA can be targeted and regulated by multiple miRNAs, and one miRNA can regulate a large number of mRNAs, simultaneously. With the exception of one miRNA that had a negative correlation with its target mRNAs, the miRNA and target mRNA expression profiles from ST cells infected with YN15 or YN144 were positively correlated. Our research also revealed DE miRNAs with target mRNAs that change in the opposite direction and form inversely correlated DE miRNA–mRNA interaction pairs, including up-regulated miRNA-down-regulated mRNA pairs and down-regulated miRNA-up-regulated mRNA pairs (Supplementary Table S11). The numbers of inversely regulated DE miRNA–mRNA pairs in PEDV-infected ST cells in the three comparison groups are shown in Table 2.
Joint functional analysis of DE miRNA and inversely DE target mRNA in PEDV-infected ST cells. (a) Flow diagram of joint analysis of DE miRNA–mRNA pairs in the three comparison groups. (b) GO analysis of DE miRNA–mRNA pairs between YN15 infection and YN144 infection. The top panel shows the GO enrichment analysis of up-regulated miRNA-down-regulated mRNA pairs, and the lower panel shows the GO enrichment analysis of down-regulated miRNA-up-regulated mRNA pairs during YN15 infection compared with YN14 infection. (c) KEGG analysis of the genes of the DE miRNA–mRNA pairs between YN15 infection and YN144 infection. The left panel shows the KEGG pathways of the top 20 significantly up-regulated miRNA-down-regulated mRNA pairs, and the right panel shows the KEGG pathways of the top 20 significantly down-regulated miRNA-up-regulated mRNA pairs during YN15 infection compared with YN144 infection. (d) The DE miRNA–mRNA interaction networks in PEDV-infected ST cells. The left panel shows the significant interactions of the up-regulated miRNA-down-regulated mRNA pairs, and the right panel shows the significant interactions of the down-regulated miRNA-up-regulated mRNA pairs during YN144 infection in comparison with YN15 infection. p-value < 0.05.
A joint analysis of the interactions between DE miRNA–mRNA pairs with a negative correlation was performed. When comparing ST cells infected with YN15 and cells infected with YN144, GO and KEGG enrichment among up-regulated and down-regulated DE target mRNAs were performed. The top 20 most significantly enriched GO terms are involved in the biological functions of bp, cc and mf (Fig. 5c). All KEGG enrichment pathways of the DE target mRNAs are shown in Supplementary Table S12. The 20 most significantly enriched KEGG mapped pathways of the inversely regulated DE miRNA–mRNA pairs are shown in Fig. 5d. These DE miRNA–mRNA pairs were jointly analyzed according to functionally enriched GO terms and enriched KEGG pathways, and the two interaction networks were formed using Cytoscape software (Fig. 5e). One interaction network shows that the differentially up-regulated miRNA-down-regulated mRNA pairs during YN15 infection in comparison with YN144 infection are involved in focal adhesion, endocytosis, regulation of action cytoskeleton, PI3K-Akt signaling pathway, and bacteria invasion of epithelial cells. The second interaction network shows that the differentially down-regulated miRNA-up-regulated mRNA pairs are involved in the NF-κB signaling pathway, TNF signaling pathway, B cell receptor signaling pathway, Toll-like receptor signaling pathway, T cell receptor signaling pathway, Janus kinase/signal transduction and activator of transcription (JAK-STAT) signaling pathway, and apoptosis.
Discussion
miRNA–mRNA interaction networks have emerged as crucial regulators in virus-host interactions, and NGS technology has become a powerful tool to analyze the expression profiles of miRNA and mRNA12. In view of the significant differences in pathogenesis and epigenetic mechanisms between the variant virulent PEDV strain YN15 and the attenuated vaccine-like PEDV strain YN144 in vivo, we suppose that miRNA and mRNA may play essential roles in PEDV virulence. This is the first reported comparative analysis of ST cell responses to infection with a virulent and attenuated strain of PEDV using small RNA-seq and mRNA-seq to explore the molecular mechanisms in the virus-host interaction.
According to our sequencing data, the ST cell transcriptional responses to infection with PEDV YN15 (virulent) and YN144 (attenuated) strains were very different in the early stage of infection (24 h pi, MOI = 0.001). According to the comprehensive GO and KEGG pathway enrichment analysis of DE miRNA–mRNA pairs with negatively correlated expression patterns in the YN15 vs YN144 comparison group, the main signaling pathways enriched were involved in host immunity and viral pathogenicity.
The main innate immune-related signaling pathways including NF-κB signaling pathway and JAK-STAT signaling pathway excited our interest. The NF-κB signaling pathway plays a crucial part in host immunity during viral infections13. The PEDV N protein is capable of inhibiting the activation of this signaling pathway activation in HEK-293 T cells14. Our sequencing data shows that NFκB1, a member of the NF-κB family of transcription factors that can activate the NF-κB signaling pathway, was substantially up-regulated in ST cells during YN144 infection (YN144 vs Control, p-value = 3.603E−31), while its expression during YN15 infection was moderate (YN15 vs Control, p-value = 5.82E−9).
The JAK-STAT signaling pathway also plays a major role in host innate immunity, especially as it pertains to inflammatory pathogenesis as seen in IBD15. In this work, cytokines related to this signaling pathway were found to be up-regulated during variant PEDV strain infection, especially during YN144 infection. The phosphorylation and translocation of STAT1, STAT2 and STAT3 promotes the antiviral ability of cells by up-regulating the transcription of interferon (IFN) -stimulated genes (ISG) to fight viral infection16. Our deep sequencing data showed that STAT1 and STAT3 were markedly up-regulated in YN144-infected ST cells (YN144 vs Control, STAT1: p-value = 9.0857E−17, and STAT3: p-value = 3.00E−11) compared with YN15-infected ST cells (YN15 vs Control, STAT1: p-value = 6.43E−12, and STAT3: p-value > 0.05). STAT2 expression displayed no obvious changes during YN144 infection (YN144 vs Control, p-value > 0.05), but was notably decreased during YN15 infection (YN15 vs Control, p-value = 6.81E−08). As key members of this signaling pathway, both the oligoadenylate synthetase (OAS) protein family (OAS1, OAS2, and OASL) and the Mx dynamin-like GTPases (MX1 and MX2) induced by type I and type III IFN participate in antiviral activity and inhibit virus replication17,18. Our research shows that the expressions of the above genes can be substantially activated at early stages of YN144 infection, while their expressions were moderate at early stages of YN15 infection (YN15 vs YN144, OAS1: p-value = 1.6782E−130, OAS2: p-value = 1.8062E−247, OASL: p-value = 3.51E−11, MX1: p-value = 3.94E−25, and MX2: p-value = 8.38E−252).
In this work, the NF-κB signaling pathway was found to be regulated by ssc-miR-155-5p, ssc-miR-29b, and ssc-miR-139-5p, and the JAK-STAT signaling pathway was regulated by ssc-miR-155-5p, ssc-miR-29b, ssc-miR-1307, ssc-miR-10a-5p, and ssc-miR-30d. All of miR-155-5p, miR-139-5p and miR-29b have been proved to have immense effects on inflammation and negatively regulate the innate immune responses to various microorganisms19,20,21. For example, miR-155 and miR-139-5p can constitute a negative feedback loop in the NF-κB signaling pathway by targeting multiple key proteins, leading to the repression of NF-κB activation in response to viral or microbial stimuli19,20. The expression of miR-155 was enhanced in microglial cells during Japanese encephalitis virus (JEV) infection, and JEV-induced IFN-β as well as downstream ISG mRNA expression was significantly reduced in microglial overexpressing miR-15519.The knock-out or inhibition of miR-155 decreases hepatitis C virus (HCV) viremia in children with leukemia22. miR-29b is capable of inhibiting the JAK-STAT signaling pathway by IFN-γ in CD8 + T cells and TH1 effector cells21. Interestingly, our sequencing data showed that ssc-miR-155-5p and ssc-miR-139-5p were markedly up-regulated in YN15-infected ST cells compared with YN144-infected ST cells (YN15 vs YN144, ssc-miR-155-5p: p-value = 4.991E-03, and ssc-miR-139-5p: p-value = 1.671E-3). Ssc-miR-29b was significantly up-regulated in ST cells infected with YN15, but down-regulated in cells infected with YN144. These differences could explain our previous results that YN144 can more effectively activate the host immune response than YN155.
A particularly noteworthy phenomenon was observed for the expression patterns of ssc-miR-1307, ssc-miR-10a-5p and ssc-miR-30d. All three were markedly down-regulated in ST cells during YN15 infection, but were significantly up-regulated during YN144 infection (shown in Supplementary Table S8). Interestingly, many studies have shown that all three play crucial roles in promoting host innate immunity during viral infection23,24,25. Qi et al. have provided evidence that miR-1307 can be significantly up-regulated in PK-15 cells after infection with foot-and-mouth disease virus23. The overexpression of this miRNA greatly up-regulates the expression of IFN-β, ISG54, 2′,5′-OAS, and NF-κB subunit p65/RELA, and activates the innate immune response, including the NF-κB and JAK-STAT signaling pathways, at early stages of viral infection23. Other research has demonstrated that miR-10a is highly expressed in the intestines and plays a crucial role in the host immune response to the microbiota in a mouse model, and the intestinal microbiota negatively regulates miR-10a expression by targeting IL-12/IL-23p4024. The miR-30 family plays an important role in the immune response during viral infection28. miR-30b and miR-30c are markedly up-regulated with IFN-α treatment and inhibit HCV replication in Huh 7.5 cells25.
Interestingly, we found that the above DE miRNAs are all involved in host immune action, although through different means and pathways. All of the DE miRNAs that were up-regulated during YN144 infection and down-regulated during YN15 infection are capable of activating the host immune response. Even more surprising, the same phenomenon occurs among all commonly up-regulated miRNAs that the expression levels of the up-regulated miRNAs during YN15 infection were significantly higher than during YN144 infection, and all of them have been reported to inhibit the NF-κB or JAK-STAT signaling pathway. Therefore, we predict that the above DE miRNAs may play an indispensable role in regulating the host innate immunity during early PEDV infection in vitro, and the above differences may be some of the reasons that YN15 and YN144 can cause different immunity action in vitro and vivo.
In contrast, YN15 can invade intestinal epithelial cells and cause serious intestinal injury and inflammation, leading to serious diarrhea in piglets. It is worth noting that diarrhea in piglet is often caused by viral-bacterial co-infections in the farm setting26. Coincidentally, the GO and KEGG enrichment analysis of up-regulated mRNA-down-regulated miRNA pairs in YN15-infected ST cells compared with YN144-infected ST cells, showed that the major signaling pathways represented include focal adhesion, regulation of action cytoskeleton, endocytosis, PI3K-Akt signaling pathway, and bacterial invasion of epithelial cells. All of these pathways are closely related to pathogenicity, and in particular, the destruction of host cell structure during microbial invasion27.
Integrin (ITG), vinculin (VCL), and fibronectin (FN1) are associated with these signaling pathways. ITGs are heterodimeric proteins expressed on the cell surface that act as receptors for the interaction between cells and microorganisms and are exploited by microorganisms to gain entry into host cells28. ITGαvβ3 is an entry receptor for PEDV infection in Vero E6 and porcine intestinal epithelial cells29. During enterohemorrhagic E. coli (EHEC) O157:H7 infection, ITGβ1 on the surface of the intestinal epithelia acts as receptors for intimin expressed by EHEC, to promote bacterial infection30.
Both talin and VCL are actin-cytoskeleton-associated, adapter proteins for the dynamic interaction between ITG and actin cytoskeleton to generate focal adhesions between cells and ECM and establish bacterial and viral infections31. For example, Ebola virus infection promotes talin-VCL expression31. IpaA secreted by Shigella interacts with VCL to initiate the formation of focal adhesion-like structures required for the efficient bacterial invasion of epithelial cells32. FN1 can polymerize to form a linear and branched meshwork on the cell surface, and a vast number of bacteria and viruses, such as Staphylococcus aureus and Influenza A Virus, express FN-binding proteins to promote cellular invasion33,34,35.
The PI3K-Akt signaling pathway is activated by variety of pathogens leading to accentuate diseases36. For example, activation of this signaling pathway promotes actin rearrangement, leading to the host inflammatory response that persists in IBD37. Additionally, cell destruction caused by viruses can expose cryptic receptors to promote bacteria adhesion and it is well known, that some symbiotic gut microbiota and invasive intestinal pathogens invade epithelial cells, and cause extensive intestinal damage38,39,40. Transmissible gastroenteritis virus (TGEV) infection can expose the adhesion proteins of intestinal epithelial cells, thereby providing the attachment sites for enterotoxigenic E. coli K88 (ETEC) invasion, inducing an epithelial–mesenchymal transition-like phenotype, and resulting in virus-bacteria co-infection41. Like TGEV, PEDV disrupts porcine intestinal epithelial cells; however, the mechanism of PEDV and bacteria co-infection is not clear40. Interestingly, paramyxovirus and Streptococcus pneumonia co-infection can only be supported by the highly pathogenic virus strains and not by the low pathogenic virus strains42. In addition, mTOR, a downstream factor of the PI3K-Akt signaling pathway, can inhibit T helper cell differentiation and thus suppress the host immune response43.
Importantly, the above phenomenon is reflected in our sequencing data demonstrationg that YN15 causes significantly greater activation of the above signaling pathways than dose YN144, as measured by the up-regulation of effector genes encoding mTOR, FN1, VCL, ITGα3, ITGα5, ITGα6, ITGα8, ITGβ1, and ITGβ4 (YN15 vs YN144, p-value = 7.98E-06, p-value = 3.27E-04, p-value = 8.96E-08, p-value = 5.25E-12, p-value = 3.27E-04, p-value = 9.61E-20, p-value = 2.04E-12, p-value = 1.41E-03, and p-value = 2.52E-12, respectively) in ST cells. This may provide an explanation as to why YN15 causes serious intestinal inflammation and pathological damage, whereas YN144 does not.
We found that several DE miRNAs regulate these pathogenicity-related signaling pathways, including ssc-miR-9, ssc-miR-30d, ssc-miR-425-5p, and ssc-miR-184. miR-9-3p has been shown to suppress cell proliferation, migration and invasion via the down-regulation of FN1, ITGβ1, ITGα544. In the present study, YN15 exhibited greater inhibition of ssc-miR-9 expression and greater activation of FN1, ITGβ1, and ITGα5 expression than the YN144 strain. These results may provide us with new insights into how to inhibit PEDV replication and secondary bacterial infection. To promote cell invasion and metastasis, miR-425-5p could activate ITGβ1expression through the suppression of suppressor of cancer cell invasion (SCAI) expression45. It is worth mentioning that miR-184 is normally found in abundance in healthy individuals; however, down-regulation of miR-184 leads to epithelial thinning and prolonged wound healing, possibly contributing to pathology46. Our sequencing data show that ST cells infected with YN15 produce a more pronounced decrease in the expression of these miRNAs than when infected with YN144 (ssc-miR-9: p-value = 3.624E-03, ssc-miR-425-5p: p-value = 7.41E-04, and ssc-miR-184: p-value = 3.92E-07). Overexpression of miR-30 in breast cancer cells inhibits the expression of a number of genes associated with invasiveness (e.g., ITGα5, ITGβ3) and tumor cell invasion in vitro47. Our sequencing data show that ssc-miR-30d were up-regulated during YN144 infection and down-regulated during YN15 infection.
In summary, PEDV infection can activate and inhibit various of genes in the same signaling pathway, and the same gene can simultaneously play different roles in different signaling pathways and present different expression ability in different stage. Based on our sequencing data, genes enrichment analysis, and information from the literature, we speculate that YN15 infection would cause more serious pathological damage, and YN144 infection would markedly activated host immune responses in vivo and vitro, which correspond well with our previous studies in piglets and prediction that YN144 does not causes clinical symptoms, but still results in effective antibody production comparable to levels achieved by vaccination5. All differences are very attractive in helping researchers unravel a new antiviral mechanism of PEDV infection, and therefore provide researchers with further avenues to the porcine industry.
Materials and methods
Cells and viruses
ST cells were purchased from American Type Cultrue Collection (ATCC) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen), supplemented with 8% Fetal Bovine Serum (BI) at 37 °C under 5% CO2. The variant virulent strain,YN1, was isolated from a sucking piglet with acute diarrhea, and was passaged to 15 generations in Vero cells, thus obtaining the virulent strain (Accession no. KT021228)5. The attenuated strain, YN144 (Accession no. KT021232), was obtained by passaging the same parent strain, YN1, to 144 generations in Vero cells. The detailed attenuation method and genomic comparison results were reported in our previous study5.
Virus infection
ST cells were cultured for approximately 24 h to 90% confluence and were washed three times with serum-free DMEM. The titers of YN15 and YN144 in ST cells were 104.1 median tissue culture infective dose (TCID50)/100 μL and 105.5 TCID50/100 μL. Three experimental groups were designed; YN15-, YN144-, and mock-infected ST cells. Each group contained three independent biological replicates, which were named Control 1, Control 2, Control 3, YN15-1, YN15-2, YN15-3, YN144-1, YN144-2 and YN144-3. ST cells were infected separately with YN15 or YN144 at a multiplicity of infection (MOI) of 0.001 for 24 h. Mock-infected ST cells were used as control. After 1 h incubation at 37 °C under 5% CO2, unbound viruses were washed and discarded three times using serum-free DMEM. The cells were then incubated with serum-free DMEM containing 8 μg/mL trypsin (Invitrogen).
Indirect immunofluorescence assay (IIFA)
ST cells infected with YN15 and YN144 PEDV strains were fixed with cold ethanol at 12, 24, and 36 h pi, respectively, and incubate with mouse monoclonal antibody against PEDV S protein for 1 h at 37 °C. Then, ST cells were washed three times with PBS before further incubation with Alexa Fluor 488-conjugated donkey anti-mouse IgG (H + L) (Thermo Fisher) in the dark. After washing five times with PBS, the images were captured under a fluorescence microscopy. Mock-infected ST cells at 36 h pi were set as control.
RNA isolation and sample preparation
Total RNA was extracted from each biological repoicate of viral-infected or mock-infected ST cells using Trizol reagent (Invitrogen) and Polyacryl Carrier (MRC) at 24 h pi. RNA degradation and contamination were monitored on 1% agarose gals. RNA purity, integrity, and concentration were measured using the NanoPhotometer spectrophotometer (IMPLEN), the RNA Nano 6000 Assay Kit of the Aglient Bioanalyzer 2100 system (Aligent Technologies) and Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies). Only RNA samples with a ratio of absorbance (260/280 nm) > 1.8 and RNA integrity numbers (RINs) > 7 were used for RNA profiling.
Library preparation and sequencing of mRNA
The library preparation and sequencing analyses of RNA were performed by Novogene Company (Bejing, China). A total quantity of 3 μg RNA per sample was used as input for the RNA sample preparations. Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) and index codes were added to attribute sequences to each sample. In order to select cDNA fragments of 150–200 bp in length, library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, USA). The PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina). The library preparations were then sequenced on an Illumina HiSeq platform and 125 bp/150 bp paired-end reads were generated. The index of the reference genome was constructed using Bowtie v2.2.3 and paired-end clean reads were aligned to the reference genome using TopHat v2.0.12. HTSeq v0.6.1 was used to count the read numbers mapped to each gene. Then, fragments per kilobase of exon model per million reads mapped (FPKM) of each gene was calculated based on the length of the gene and numbers of reads mapped to the gene48.
Library preparation, sequencing, prediction and analysis of miRNA
A total of 3 μg RNA per sample was used as input for each small RNA library. NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA) was used to generate the sequencing libraries and index codes that were added to attribute sequences to each sample. Library quality was assessed on the Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips. After cluster generation of the index-coded samples, performed on a cBot Cluster Generation System using TruSeq SR Cluster Kit v3-cBot-HS (Illumina), the library was sequenced on an Illumina HiSeq 2500/2000 platform, and 50 bp single-end reads were generated.
The small RNA tags were mapped to the Sus scrofa sequence by Bowtie49. The miRBase20.0 database was used as the reference, and modified software mirdeep2 and sRNA-tools-cli were used to obtain potential miRNA and predict their secondary structures50. The available software miREvo and mirdeep2 were integrated to predict novel miRNAs50,51.
To explore the occurrence of the miRNA families identified in this study in other species, miFam.dat (http://mirbase.org/ftp.shtml) was used to search for known miRNA families, and novel miRNA precursors were submitted to Rfam (http://rfam.sanger.ac.uk/search/) to search for Rfam families.
miRNA expression levels were estimated by TPM (transcript per million) using the following criterion- normalization formula: normalized expression = mapped read count/ total reads × 100000052.
Differential expression of miRNA and mRNA, and target gene prediction
Analysis of DE miRNA from the three experimental groups (YN15, YN144 and control) was performed using the DESeq R package (1.8.3). Differentially expressed mRNAs from the three comparison groups were analyzed using the DESeq R package (1.18.0). The p-value was adjusted using the Benjamini & Hochberg method. A corrected p-value of 0.05 was set as the threshold for significantly different expression by default (p-value < 0.05, and padj < 0.05). Prediction of the miRNA target gene was performed using psRobot-tar in miRanda for Sus scrofa53.
GO and KEGG enrichment analysis of differentially expressed genes
Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs were performed using Goseq R package and KOBAS 2.0 software (Available online: http://kobas.cbi.pku.edu.cn/home.do).
Correlation analysis of DE miRNAs and DE mRNAs
To generate the miRNA–mRNA interaction network, a negative correlation analysis of DE miRNAs and DE mRNAs was performed, and Cytoscape v2.8.3 software (http://www.cytoscape.org/) was used to construct the miRNA–mRNA interaction network.
Real-time relative quantitative reverse transcription polymerase chain reaction (qRT-PCR)
To validate the sequencing data, 6 DE mRNAs and 6 DE miRNAs were randomly selected and analyzed using real-time relative qRT-PCR and three independent biological replicates were performed at minimum.
For detection of DE mRNAs, RNA was reverse transcribed into single strand cDNA using Prime Script RT reagent Kit RR036A (TaKaRa, Shiga-ken, Japan) on a S1000 Thermal Cycler (Bio-Rad, CA, USA). The GAPDH gene was used as the internal control. The primers used are listed in Supplementary Table S13.
For detection of DE miRNAs, RNA was reverse transcribed into single strand cDNA using PrimeScript RT reagent Kit RR037A (TaKaRa, Shiga-ken, Japan) on a S1000 Thermal Cycler (Bio-Rad, CA, USA). Hieff qPCR SYBR Green Master Mix (Low Rox Plus) (YEASEN, Shanghai, CHN) was used to perform qRT-PCR. The expression of U6 small nuclear RNA (snRNA) was used as the endogenous control. The primers used are listed in Supplementary Table S13.
Real-time relative qRT-PCR was performed in a 10 μL volume containing 100 nM of each of forward primer and reverse primer, cDNA template, and Faststart DNA Master SYBR Green I Mix reagent kit (Roche) on a ViiA 7 Real-Time PCR System (Life Technologies, CA, USA) under the following cycle conditions: 95 °C for 10 min, 40 cycles with 95 °C for 15 s, 56 °C for 30 s, and 72 °C for 31 s.
The relative expression of each target was calculated using the 2-ΔΔCT method with a control group as calibrator54. Statistical significance was determined by Student’s t-test, with p-value < 0.05 deemed to be statistically significant.
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Code availability
Any custom code used during the current study is available from the corresponding author on reasonable request.
References
Pensaert, M. B. & de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 58, 243–247 (1978).
Fang, C., Xu, K. & Zhong, L. Genetic characteristics of porcine epidemic diarrhea virus in Chinese mainland, revealing genetic markers of classical and variant virulent parental/attenuated strains. Gene 588, 95–102 (2016).
Masuda, T., Murakami, S. & Takahashi, O. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 160, 2565–2568 (2015).
Huang, Y. W., Allan, W. D. & Pablo, P. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4, e00737 (2013).
Fang, C., Yin, X. & Mei, W. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses 7, 5525–5538 (2015).
Zhong, L., Fang, C. & Shi, Y. Comparative proteome analysis of porcine jejunum tissues in response to a virulent strain of porcine epidemic diarrhea virus and its attenuated strain. Viruses 8, 323 (2016).
Yasukawa, K., Kinoshita, D. & Nakagawa, T. The microRNAs miR-302b and miR-372 regulate mitochondrial metabolism via the SLC25A12 transporter, which controls MAVS-mediated antiviral innate immunity. J. Biol. Chem. 295, 444–457 (2020).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Li, L., Fei, G. & Yi, J. Cellular miR-130b inhibits replication of porcine reproductive and respiratory syndrome virus in vitro and in vivo. Sci. Rep. 5, 17010 (2015).
Li, L., Zu, W. & Yan, Z. Host miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by upregulating type I interferons. Virus Res. 195, 86–94 (2015).
Gupta, A., Pruthvi, N. & Hai, L. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1). PLoS ONE 6, e22730 (2011).
Ma, X., Becker Buscaglia, L. E. & Barker, J. R. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 3, 159–166 (2011).
Li, C., Xu, G. & Yu, G. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 96, 1757–1767 (2015).
Coskun, M., Salem, M. & Pedersen, J. Involvement of JAK/STAT signaling in the pathogenesis of inflammatory bowel disease. Pharmacol. Res. 76, 1–8 (2013).
Mitchell, T. J. & John, S. Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas. Immunology 114, 301–312 (2005).
Kristiansen, H., Gad, H. H. & Eskildsen-Larsen, S. The oligoadenylate synthetase family: An ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 31, 41–47 (2011).
Haller, O., Staeheli, P. & Schwemmle, M. Mx GTPases: Dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23, 154–163 (2015).
Pareek, S., Roy, S. & Kumari, B. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflammation 11, 97 (2014).
Ming, Z., Wen, Z. & Jun, M. MicroRNA-139-5p regulates chronic inflammation by suppressing nuclear factor-κB activity to inhibit cell proliferation and invasion in colorectal cancer. Exp. Ther. Med. 18, 4049–4057 (2019).
Gigante, M., Paola, P. & Wolfgang, H. miR-29b and miR-198 overexpression in CD8+ T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J. Transl. Med. 14, 84 (2016).
Hassan, S. S., Nashwa, E. & Elshimy, A. A. In vitro knock-out of miR-155 suppresses leukemic and HCV virus loads in pediatric HCV-4–associated acute lymphoid leukemia: A promising target therapy. J. Cell. Biochem. 121, 2811–2817 (2020).
Qi, L., Wang, K. & Chen, H. Host microRNA miR-1307 suppresses foot-and-mouth disease virus replication by promoting VP3 degradation and enhancing innate immune response. Virology 535, 162–170 (2019).
Xue, X., Feng, T. & Yao, S. Microbiota downregulates dendritic cell expression of miR-10a, which targets IL-12/IL-23p40. J. Immunol. 187, 5879–5886 (2011).
Zhang, X., Daucher, M. & Armistead, D. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS ONE 8, e55733 (2013).
Wang, X., Ren, W. & Ying, N. A novel watery diarrhoea caused by the co-infection of neonatal piglets with Clostridium perfringens type A and Escherichia coli (K88, 987P). Vet. J. 197, 812–816 (2013).
Elbahesh, H., Cline, T. & Baranovich, T. Novel roles of focal adhesion kinase in cytoplasmic entry and replication of influenza A viruses. J. Virol. 88, 6714–6728 (2014).
Dupuy, A. G. & Caron, E. Integrin-dependent phagocytosis: Spreading from microadhesion to new concepts. J. Cell Sci. 121, 1773–1783 (2008).
Li, C., Su, M. & Yin, B. Integrin αvβ3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells. Vet. Microbiol. 237, 108400 (2019).
Sinclair, J. F., Dean-Nystrom, E. A. & O’Brien, A. D. The established intimin receptor Tir and the putative eucaryotic intimin receptors nucleolin and beta1 integrin localize at or near the site of enterohemorrhagic Escherichia coli O157:H7 adherence to enterocytes in vivo. Infect. Immun. 74, 1255–1265 (2006).
Liu, Y., Xiao, J. & Zhang, B. Increased talin-vinculin spatial proximities in livers in response to spotted fever group rickettsial and Ebola virus infections. Lab. Invest. 100, 1030–1041 (2020).
Van Nhieu, G. T. & Ben-Ze’ev, A. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMOB J. 16, 2717–2729 (1997).
Henderson, B., Nair, S. & Pallas, J. Fibronectin: A multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 35, 147–200 (2011).
Menzies, B. E. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr. Opin. Infect. Dis. 16, 225–229 (2003).
Leung, H. S., Li, O. T. & Chan, R. W. Entry of influenza A Virus with a α2, 6-linked sialic acid binding preference requires host fibronectin. J. Virol. 86, 10704–10713 (2012).
Oliveira, J. S. S., Santos, G. S. & Moraes, J. A. Reactive oxygen species generation mediated by NADPH oxidase and PI3K/Akt pathways contribute to invasion of Streptococcus agalactiae in human endothelial cells. Mem. Inst. Oswaldo Cruz. 113, e140421 (2018).
Long, S. H., Chen, M. H. & Gao, K. Activation of PI3K/Akt/mTOR signaling pathway triggered by PTEN downregulation in the pathogenesis of Crohn’s disease. J. Dig. Dis. 14, 662–669 (2013).
Peltola, V. T., Murti, K. G. & McCullers, J. A. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 192, 249–257 (2005).
Lee, I. K., Kye, Y. C. & Kim, G. Stress, nutrition, and intestinal immune responses in pigs—a review. Asian-Austr. J. Anim. Sci. 29, 1075–1082 (2016).
Cao, L., Ge, X. & Ren, Y. Porcine epidemic diarrhea virus infection induces NF-κB activation through the TLR2, TLR3 and TLR9 pathways in porcine intestinal epithelial cells. J. Gen. Virol. 96, 1757–1767 (2015).
Arango Duque, G. & Acevedo Ospina, H. A. Understanding TGEV–ETEC coinfection through the lens of proteomics: A tale of porcine diarrhea. Proteom. Clin. Appl. 12, 1700143 (2018).
Alymova, I. V., Portner, A. & Takimoto, T. The novel parainfluenza virus hemagglutinin-neuraminidase inhibitor BCX 2798 prevents lethal synergism between a paramyxovirus and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 49, 398–405 (2005).
Essig, K., Hu, D. & Guimaraes, J. Roquin suppresses the PI3K-mTOR signaling pathway to inhibit T helper cell differentiation and conversion of Treg to Tfr cells. Immunity 47, 1067–1082 (2017).
Ding, Y., Pan, Y. & Liu, S. Elevation of MiR-9–3p suppresses the epithelial–mesenchymal transition of nasopharyngeal carcinoma cells via down-regulating FN1, ITGB1 and ITGAV. Cancer Biol. Ther. 18, 414–424 (2017).
Fang, F., Song, T. & Zhang, T. MiR-425-5p promotes invasion and metastasis of hepatocellular carcinoma cells through SCAI-mediated dysregulation of multiple signaling pathways. Oncotarget 8, 31745–31757 (2017).
Derrick, T., Roberts, C. & Rajasekhar, M. Conjunctival MicroRNA expression in inflammatory trachomatous scarring. PLoS Negl. Trop. Dis. 7, e2117 (2013).
Croset, M., Pantano, F. & Kan, C. W. S. miRNA-30 family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Cancer Res. 78, 5259–5273 (2018).
Trapnell, C., Williams, B. A. & Pertea, G. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Langmead, B., Trapnell, C. & Pop, M. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Friedländer, M. R., Mackowiak, S. D. & Li, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012).
Wen, M., Shen, Y. & Shi, S. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 13, 140 (2012).
Zhou, L., Chen, J. & Li, Z. Integrated profiling of MicroRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 5, e15224 (2010).
Wu, H. J., Ma, Y. K. & Chen, T. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40, W22–W28 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
This work was granted by Natural Science Foundation of China (No. 31972667).
Author information
Authors and Affiliations
Contributions
X.Z. has full access to all of the data in the research and takes responsibility for the integrity of the data and the accuracy of the data analysis; Research conception and design: X.Z., C.L., B.Z., Z.L., W.Z., R.L., G.C., J.C., S.F., and Q.H.; Acquisition of data: X.Z., C.L., B.Z., Z.L., and W.Z.; Analysis and interpretation of data: X.Z., C.L., B.Z., R.L., G.C., J.C., and S.F.; X.Z. wrote the paper. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, X., Li, C., Zhang, B. et al. Differential expression and correlation analysis of miRNA–mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci Rep 11, 1868 (2021). https://doi.org/10.1038/s41598-021-81189-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-81189-5
This article is cited by
-
Integrated analysis of microRNA and messenger RNA expression profiles reveals functional microRNA in infectious bovine rhinotracheitis virus-induced mitochondrial damage in Madin-Darby bovine kidney cells
BMC Genomics (2024)
-
Coexpression analysis of lncRNAs and mRNAs identifies potential regulatory long noncoding RNAs involved in the inflammatory effects of lipopolysaccharide on bovine mammary epithelial cells
BMC Veterinary Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.