Abstract
Palustrin-Ca (GFLDIIKDTGKEFAVKILNNLKCKLAGGCPP) is a host defence peptide with potent antimicrobial and anticancer activities, first isolated from the skin of the American bullfrog Lithobates catesbeianus. The peptide is 31 amino acid residues long, cationic and amphipathic. Two-dimensional NMR spectroscopy was employed to characterise its three-dimensional structure in a 50/50% water/2,2,2-trifluoroethanol-\(d_{3}\) mixture. The structure is defined by an \(\alpha\)-helix that spans between Ile\(^{6}\)-Ala\(^{26}\), and a cyclic disulfide-bridged domain at the C-terminal end of the peptide sequence, between residues 23 and 29. A molecular dynamics simulation was employed to model the peptide’s interactions with sodium dodecyl sulfate micelles, a widely used bacterial membrane-mimicking environment. Throughout the simulation, the peptide was found to maintain its \(\alpha\)-helical conformation between residues Ile\(^{6}\)-Ala\(^{26}\), while adopting a position parallel to the surface to micelle, which is energetically-favourable due to many hydrophobic and electrostatic contacts with the micelle.
Similar content being viewed by others
Introduction
Host defence peptides (HDPs) are naturally occurring molecules that are secreted as part of the non-specific response of the innate immune system1. These molecules contribute to the immune system’s antimicrobial response either by exerting direct antimicrobial activity, or by altering the host’s immune response. HDPs are particularly prevalent in amphibians, with approximately one-third of all known HDPs having been originally isolated from amphibians. Nonetheless, HDPs are a highly conserved part of the immune response, and have been found in all species of organisms, from humans to bacteria2,3,4,5.
As the number of documented cases of antimicrobial resistance to conventional small-molecule drugs continues to rise, interest in antimicrobial peptides (AMPs), which can exert their antimicrobial activity against both Gram-positive bacteria and Gram-negative bacteria without the development of resistance, continues to grow6. AMPs possess a number of additional advantages. They are more efficacious, selective and specific than small molecules, and are degraded to naturally occurring amino acids, which reduces the danger of unfavourable drug-drug interactions. AMPs are suitable for use both in combination with conventional drugs, and as standalone substitutes. The biological activities of HDPs, however, are not limited to antimicrobial activity. HDPs have also been demonstrated to possess antifungal, antiparasitic, antiviral and anticancer activities7,8,9,10. Indeed, palustrin-Ca (GFLDIIKDTGKEFAVKILNNLKCKLAGGCPP) is reported to be the most potent known anticancer peptide, with an IC\(_{50}\) of 0.951 μg/ml against human gastric cancer SGC-790111.
Typically, HDPs are short, with a primary sequence length between 10 and 50 amino acid residues. Usually, HDPs are unstructured in pure aqueous solution, and only adopt a defined secondary structure upon contact with a biological membrane, or in a hydrophobic environment. The induced structures are predominantly \(\alpha\)-helical in nature, although other identified structures include \(\beta\)-sheet, mixed \(\alpha\)-\(\beta\) and extended structures. A common feature of HDP structures is that they are typically amphipathic in nature, with a clear separation between the peptide’s hydrophobic and polar or charged residues. Overall, HDP’s are usually positively charged, although a number of negatively charged peptides have been isolated as well12. Indeed, increased positive charge, up to a certain extent, has been associated with greater antimicrobial activity13,14. Similarly, increased peptide hydrophobicity is associated with peptide haemolytic activity, and thereby represents another key feature that can be changed to modulate peptide selectivity15,16. Interestingly, peptide dimerisation has emerged as a potential strategy for increasing AMP effectiveness by enhancing pore formation17,18.
While HDPs are typically positively charged, anionic molecules such as lipopolysaccharides (LPS), phospholipids and teichoic acids are prevalent in prokaryotic cell membranes, which enables attractive electrostatic interactions between HDPs and their target bacterial cell membranes19,20. While peptide cationicity is generally a requirement for attraction to membranes, the loss of N-terminal positive charge has been shown to allow a deeper peptide insertion into the lipid bilayer21, although a separate study has found that reduced N-terminal cationicity results in reduced antimicrobial activity22. Similarly to prokaryotic membranes, the cell membranes of cancer cells are enriched in negative charge, unlike non-cancerous cell membranes which are neutral, which allows anticancer peptides to be selectively electrostatically attracted to cancer cells.
HDPs usually exert their bactericidal activity via disruption of the bacterial cell membrane, for which three main mechanisms of action have been described: the toroidal pore model, barrel-stave model and carpet model. All three membranolytic mechanisms of action are relatively non-specific, which is the reason why few cases of bacterial resistance to HDPs have been described23,24. The initial step in all three mechanisms is the HDP binding of the target membrane, initially facilitated via the aforementioned electrostatic attraction, then via hydrophobic attraction. As the peptide concentration at the membrane increases, eventually a threshold concentration is reached, whereupon the peptides begin to induce changes in the membrane structure25. As the peptide concentration continues to increase, a second threshold concentration is reached, at which membrane lysis occurs25. This concentration may be determined by the organism’s capacity to repair its damaged cell membranes26.
Generally, membrane lysis occurs either via the pore-forming toroidal pore or barrel-stave models, or the non-pore-forming carpet model. The latter requires that the bilayer curvature is disrupted, thereby disintegrating the bacterial membrane. Under the toroidal pore model, the bacterial membrane is bent from the outer leaflet inwards, resulting in the formation of a transmembrane toroidal pore lined by the peptides and the membrane’s lipid head groups27,28. The barrel-stave model, meanwhile, involves the peptides forming a transmembrane pore; its lumen is lined by the peptides’ hydrophobic residues, and its exterior is composed of hydrophilic residues, which are in contact with the membrane’s lipid headgroups.
Where an amphipathic peptide structure is modified so that a residue side chain on its hydrophobic face has switched polarity, membrane lysis can also occur via a fractal rupture mechanism. This mechanism relies on apertures in the upper leaflet of the lipid bilayer being caused by chemically induced peptide-lipid interfaces, thereby exposing the membrane’s hydrophobic layer and resulting in an energetically unfavourable water-lipid interface, and ultimately in structured defects in the membrane29.
Interestingly, some HDPs exert their activity without disruption of the lipid bilayer. Instead, these peptides translocate across the membrane without causing lysis, and instead exert antimicrobial activity by targetting an intracellular target30.
Our laboratory has conducted structural studies on various amphibian HDPs, including ranatuerin-2CSa31, XT-732, alyteserin-1c33, brevinin-1BYa34 and its analogues35, maximin 336 and maximin 137. Palustrin-Ca (GFLDIIKDTGKEFAVKILNNLKCKLAGGCPP) is a 31 amino acid residue HDP first isolated from the skin of the American bullfrog Lithobates catesbeianus11. In common with other HDPs isolated from frogs belonging to the Ranidae family, it contains a cyclic disulfide bridged domain at the C-terminal end of the peptide sequence, between residues 23 and 29. Palustrin-Ca is biologically interesting, as it is non-haemolytic, and exhibits potent broad-spectrum antibacterial activity, and as previously mentioned, very potent anticancer activity, with an IC\(_{50}\) of 0.951 μg/ml against human gastric cancer SGC-790111. In this work, palustrin-Ca’s conformation and structural properties in a 50% H\(_{2}\)O-trifluoroethanol-\(d_{3}\) mixture are elucidated using NMR spectroscopy, with an ensemble of model structures being determined. Additionally, the peptide’s interactions with a membrane-mimetic are simulated using an atomistic molecular dynamics simulation.
Materials and methods
Materials and NMR sample preparation
The palustrin-Ca peptide (MW = 3304 g/mol, purity > 95%) was purchased from ProteoGenix (Paris). The identity and purity of the peptide were confirmed using mass spectrometry and high performance liquid chromatography (Supplementary Information). 4.5 g of the peptide was dissolved in 0.6 mL of a 50% (v/v) TFE-\(d_{3}\)/H2O solution, resulting in a final peptide concentration of 2.27 mM. 3-trimethylsilyl propionic acid (TSP) and 2,2,2-trifluoroethanol (TFE-\(d_{3}\)) of analytical grade were obtained from Sigma-Aldrich (Ireland).
NMR spectroscopy
A Bruker Avance 600 NMR spectrometer with a 5 mm inverse probe head at a 1H resonance frequency of 600.13 MHz was used to perform a number of NMR experiments at a temperature of 298 K. One-dimensional proton, 2D phase-sensitive total correlation spectroscopy (TOCSY)38, nuclear Overhauser effect spectroscopy (NOESY)39 and natural abundance \(^{1}\)H-\(^{13}\)C and \(^{1}\)H-\(^{15}\)N heteronuclear single quantum coherence spectroscopy (\(^{1}\)H-\(^{13}\)C-HSQC) and \(^{1}\)H-\(^{15}\)N-HSQC40 spectra were acquired, with relaxation delays of 2.5 s, 2.0 s, 1.5 s, 1.0 s and 2.0 s, respectively, and acquisition times of 3.4 s, 340 ms, 280 ms, 170 ms and 100 ms, respectively. Mixing times of 60 ms and 200 ms were used for the TOCSY and NOESY, respectively.
The 1H spectral widths were 6.0 kHz, 7.2 kHz, 6.0 kHz and 9.6 kHz, and the spectra were acquired with 4, 16, 32 and 64 transients for each of the 1024, 2048, 256 and 128 t1 increments for the TOCSY, NOESY, \(^{1}\)H-\(^{13}\)C-HSQC and \(^{1}\)H-\(^{15}\)N-HSQC, respectively. The 13C spectral width was 21.1 kHz and the 15N spectral width was 10.9 kHz. All two-dimensional spectra were processed with the Bruker TopSpin program, version 4.0.6 (Bruker BioSpin, Germany), using the sine squared window function, and the 1H signal of TSP was used as the chemical shift reference.
Structure calculation
The NMRFAM-SPARKY program, version 3.13141 was used to analyse the acquired NMR spectra and integrate the NOESY cross-peaks. The integrated peak volumes were exported, and the nuclear Overhauser effect (NOE) cross-peak intensities were calibrated using the CALIBA42 program, yielding a set of upper distance restraints. Protons that could not be stereospecifically assigned were treated as pseudoatoms. The set of obtained distance restraints was used as input to CYANA43, excluding those that represent fixed distances. A force constant of 1 kJ mol\(^{-1}\) Å\(^{-2}\) was used to weight the distance restraints, from which one hundred structures were generated by CYANA44, and submitted to 20,000 steps of simulated annealing and 20,000 of conjugate gradient minimization. The 20 structures with the lowest target function values were selected, and subjected to an additional 2000 steps of conjugate gradient energy minimization with restrained backbone atoms, using the CHARMM22 force field45 in NAMD, version 2.1246. VMD (visual molecular dynamics), version 1.9.347, was used to analyse the final ensemble of 20 structures. The geometry, stereochemical quality and structural statistics of the final ensemble of structures was assessed and validated using PROCHECK48 and the wwPDB web service49.
Molecular dynamics simulation
Structural model coordinates of an SDS micelle were obtained from Jakobtorweihen et al.50, and used in the construction of an SDS micelle-peptide system, which was constructed by aligning the centres of mass of the micelle and the medoid palustrin-Ca structure with VMD. To ensure that the system’s charge remained neutral, chloride ions were added as counter-ions, and finally, TIP3P water was used to solvate the system.
The system was energy minimized and equilibrated using the CHARMM22 all-atom forcefield45,51 within NAMD version 2.1246. All calculations were conducted within the NPT ensemble, using the Langevin piston Nose-Hoover method52,53 and periodic boundary conditions. Long-range non-bonded interactions were calculated up to a switching distance of 8.5 Å, beyond which a smooth switching function truncated the energy to a cut-off of 11 Å. The PME method54 was used to calculate long-range electrostatic interactions at each time step. The non-bonded interaction list was also updated every step. All bonds to hydrogen atoms were constrained using the SHAKE algorithm55. A 2 fs timestep was employed.
The solvated peptide-micelle system was minimized for 2000 conjugate gradient steps with peptide backbone atoms fixed, and a further 2000 steps with its \(\alpha\)-carbon atoms restrained. The system was then heated to a temperature of 310 K over 6000 steps using Langevin dynamics, with \(\alpha\)-carbon remaining restrained. The system volume was equilibrated with the Langevin piston at 1 atm over 24,000 steps, with a further 24,000 steps without restraints. Finally, the system was simulated for 57.4 ns.
The radial distribution function g(r) was used for analysing the final 22.5 ns of simulation data. The measure gofr command within VMD was used to calculate the g(r) function between each residue’s carbon atoms and the SDS micelle’s non-sulfate atoms. The default values 0.1 and 25.0 Å were used for \(\delta r\) and max r, respectively.
Results and discussion
Conformational analysis by NMR
Two-dimensional NMR spectroscopy was used to determine the structure of palustrin-Ca in a 50:50 TFE-\({d_{3}}\)/water solvent mixture, which is a standard solvent mixture employed in the characterisation of peptide structures that promotes the formation of secondary structures, including \(\alpha\)-helices56. Structural studies of another ranid frog-derived HDP, brevinin-1BYa, and its analogues in TFE and SDS micelles have shown that the peptide conformations in both media are comparable34,35. The effects that TFE exerts on peptide structures are well-studied, having been the subject of research for approximately 60 years57,58.
Peptides unrestrained by disulfide bonds typically are unstructured in pure aqueous solution59,60,61,62, as \(\alpha\)-helical self-aggregation is hindered by the presence of cationic or sterically bulky residues. Furthermore, sequence enrichment in lysine, which is common among AMPs, can preclude the formation of \(\beta\)-sheets. Consequently, most linear cationic HDPs do not possess a defined secondary structure in pure aqueous solution63,64.
Nonetheless, the addition of TFE has been demonstrated to induce the formation of secondary structure by enhancing \(\alpha\)-helical character in peptides65. This has been shown in studies that systematically increase TFE concentration34,35,59,66.
It is generally understood that the mechanism by which peptide \(\alpha\)-helical character is enhanced commences with the preferential aggregation of TFE molecules around the peptide, facilitated by hydrophobic interaction between peptide’s hydrophobic sidechains and the trifluoromethyl group, which allows the TFE molecules to substitute the peptide’s hydration shell. This replacement removes alternative hydrogen-bonding partners, and results in a low dielectric environment, comparable to that experienced by peptides in biological membranes, which favours the formation of intramolecular hydrogen bonds between the backbone amide groups67,68.
NMR conformational studies of magainin 2 demonstrated that in a pure aqueous solution, the peptide possesses an extended random coil structure69. Similarly, the maximin 4 antimicrobial peptide does not have a well-defined structure in pure aqueous solution70. Previously, a 50% TFE concentration has been found to be appropriate for the characterisation of peptide structures, as low TFE concentrations result in poor amide chemical shift dispersion, which indicates a lack of defined secondary structure71.
The collected spectra were well-resolved, with well-dispersed peaks that facilitated unambiguous resonance assignment. Individual residue spin systems were identified with the aid of TOCSY, \(^{1}\)H-\(^{13}\)C HSQC and \(^{1}\)H-\(^{15}\)N HSQC spectra, while sequence-specific resonance assignment was conducted using the HN-HN and HN-H\(\alpha\) regions of the NOESY spectrum (Fig. 1). The first sequence-specific resonance assignments were made for the unique residues Thr\(^{9}\), Glu\(^{12}\) and Val\(^{15}\), with the remainder being assigned through the use of a backbone-walk, and investigation of HN-H\(\alpha\) region peaks, where necessary. The Gly\(^{1}\) amide proton chemical shift could not be determined due to chemical exchange. The presence of a disulfide bridge was confirmed by observation of clear long-range \(\beta _{i}\)-\(\beta _{i+6}\) NOE peaks between residues Cys\(^{23}\)-Cys\(^{29}\).
All the \(^{1}\)H chemical shifts identified for palustrin-Ca are detailed in Table 1. Likewise, all of palustrin-Ca’s intermolecular NOE connectivities are summarised in Fig. 2, where the NOE intensity is proportional to the thickness of each line. Multiple d\(_{\alpha}\)N(i, i+3), d\(_{\alpha \beta }\)(i, i+3), d\(_{\alpha }\)N(i, i+4) and d\(_{\alpha \beta }\)(i, i+4) were identified, which are indicative of an \(\alpha\)-helical secondary structure.
Chemical shift analysis
Amide proton chemical shift deviations, calculated as the difference between the observed chemical shift and the random coil chemical shift (\(\Delta \delta = \delta _{obs} - \delta _{rc}\)), are connected with the hydrogen bond length72. Shorter hydrogen bond lengths are associated with higher \(\Delta \delta\) values, and vice versa. Calculated amide proton chemical shift deviations are illustrated in Fig. 3A; the corresponding hydrogen bond lengths are given in Fig. 3B. The amide proton chemical shift deviations exhibit a periodicity of 3–5 from residue 8 on, with greater frequency in the C-terminal part of the sequence. The greatest values are found for the Phe\(^{2}\), Asp\(^{8}\), Phe\(^{13}\), Leu\(^{18}\), Leu\(^{21}\), Leu\(^{25}\) and Gly\(^{28}\), while the lowest values are observed for Asp\(^{4}\), Lys\(^{11}\), Lys\(^{16}\), Asn\(^{20}\), Cys\(^{23}\) and Gly\(^{27}\). As expected, the greatest values of \(\Delta \delta\) are observed for hydrophobic residues, with the exception of the Asp\(^{8}\) and Gly\(^{28}\) residues, which represent the beginning and end of the observed chemical shift deviation periodicity. Similarly, the lowest values are observed for polar and charged residues.
(A) Amide proton chemical shift deviation plot of palustrin-Ca in 50% TFE-\({d_{3}}\)-H\(_{2}\)O. The observed amide proton chemical shifts were compared to the standard random coil chemical shifts (\(\Delta \delta =\delta _{obs}-\delta _{rc}\)). (B) Hydrogen bond distance as calculated from the chemical shift deviations (\(\Delta \delta = 19.2d_{N}^{-3}-2.3\)), where \(d_{N}\) denotes hydrogen bond length72.
Similarly, \(\alpha\)-proton chemical shift deviations were calculated as the difference between the observed chemical shift and the random coil chemical shift (\(\Delta \delta = \delta _{obs} - \delta _{rc}\)). Four sequential \(\alpha\)-proton chemical shift deviations at least 0.1 ppm lower than the expected random coil values indicate \(\alpha\)-helical segments, while three or more deviations at least 0.1 ppm higher are indicative of \(\beta\)-strands; no change greater than 0.1 ppm is indicative of coiled regions73. Figure 4 shows the chemical shift deviations of palustrin-Ca, which shows a clear stretch of downfield shifts between residues Phe\(^{2}\)-Asp\(^{8}\), and again between residues Lys\(^{11}\)-Leu\(^{25}\). While these chemical shift deviations are indicative of \(\alpha\)-helical structure, the former stretch of downfield shifts may instead be attributed to a turn conformation. While the \(\alpha\)-proton chemical shift deviations for residues Ala\(^{26}\)-Pro\(^{30}\) are generally upfield, this trend is not sufficiently consistent to attribute any secondary structure to it.
Molecular modelling
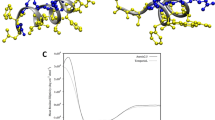
The NOESY cross-peaks were integrated, and the resultant volumes were converted into distance restraints. CYANA was employed to generate one hundred structures, of which the twenty possessing the lowest target function values were further energy-minimized. Table 2 summarizes the structural and energetic statistics of the twenty selected models. As depicted in Fig. 5, palustrin-Ca is predominantly \(\alpha\)-helical in TFE-\({d_{3}}\), which is known to promote the formation of the secondary structure. The ensemble’s secondary structural properties were analysed with the aid of STRIDE74. Most of the ensemble’s structures are display \(\alpha\)-helical structures between residues Ile\(^{6}\)-Ala\(^{26}\). The terminal segments are predominantly defined as turns, but possess significant coil character as well. More specifically, the N-terminus exhibits mostly turn character, although the coil character of the C-terminus is greater. The peptide structure is amphipathic, with a clear separation between the polar and non-polar faces of the helix. This is unsurprising, considering that amphipathicity is a common requirement for antimicrobial activity75.
Ribbon representation of the medoid solution structure of palustrin-Ca in 50% TFE-\({d_{3}}\)-H\(_{2}\)O. The amphipathic helical motif is observed. Positively charged residues are coloured in cyan, negatively charged residues are coloured in pink, polar residues are coloured in purple, glycine residues are coloured in orange, cysteine residues are coloured in green, hydrophobic residues are coloured in slate.
Molecular dynamics
A molecular dynamics simulation of palustrin-Ca’s medoid structure in an SDS micelle was conducted to characterise its behaviour in the prokaryotic membrane-mimetic environment. SDS micelles are often used as mimetics of the bacterial membrane in molecular dynamics simulations76,77,78,79,80. In the course of the simulation, the peptide translocated from its initial position aligned with the micelle’s centre of mass, to the micelle’s surface-water boundary, where it adopted a position parallel to the micelle surface (Fig. 6). The peptide’s amphipathicity is clearly apparent; its hydrophobic residues remain closely associated with the micellar interior, while the hydrophilic residues are in contact with the aqueous solvent and the micelle’s anionic sulfate headgroups. This amphipathic configuration is quite energetically favourable, and is contributes to the peptide’s exertion of its antimicrobial activity81. The peptide’s cationic residues are important for its bioactivity, as the electrostatic interaction between the target surface’s anionic charges and a peptide’s cationic residues drives the initial attraction of the peptide to the target82. Likewise, the hydrophobic residues are necessary for the peptide’s insertion into the target hydrophobic core83.
Palustrin-Ca positioned at the SDS micelle-water boundary. Lysine residue side chains are shown interacting with the negatively charged sulfate headgroups. Positively charged residues are coloured in cyan, negatively charged residues are coloured in pink, polar residues are coloured in purple, glycine residues are coloured in orange, cysteine residues are coloured in green, hydrophobic residues are coloured in slate. SDS aliphatic chains are coloured in white, while the SDS headgroups are orange and red, and highlighted with dotted spheres.
The peptide’s interactions with the micelle’s hydrophobic core were quantified using the radial distribution function g(r), which was calculated between the peptide’s carbon atoms and the micelle’s aliphatic chain, and plotted against the radius r, as shown in Fig. 7.
The radial distribution function plots illustrate clearly that the peptide’s hydrophobic residues maintain a close association with the micelle, especially in the structured \(\alpha\)-helical segment, where the hydrophobic Ile\(^{6}\), Phe\(^{13}\), Ala\(^{14}\), Ile\(^{17}\), Leu\(^{18}\), Leu\(^{21}\) and Leu\(^{25}\) residues are particularly closely associated to the micelle hydrophobic core. Conversely, hydrophilic residues, including Asp\(^{8}\), Glu\(^{12}\), Lys\(^{16}\), Asn\(^{19}\) and Lys\(^{22}\) are less associated with the micelle core, and in close contact with the aqueous solvent.
Interestingly, the N-terminal segment of the peptide is also found to maintain close association with the micelle, especially through the Gly\(^{1}\), Phe\(^{2}\) and Ile\(^{6}\) residues, despite the first five residues not forming part of a defined secondary structure. This is in contrast to the C-terminal segment, where beginning with Ala\(^{26}\) onwards the peptide is only weakly associated with the micelle.
The peptide, which was experimentally determined to be \(\alpha\)-helical between residues Ile\(^{6}\) and Ala\(^{26}\), maintains this structure throughout the duration of the simulation. Despite the presence of a glycine residue at position 10, the \(\alpha\)-helix remains relatively rigid, not exhibiting any significant flexibility during the simulation. This suggests that the glycine residue occurs too far from the helical segment’s middle to impart flexibility. Although studies have previously demonstrated that a rigid \(\alpha\)-helical structure can potentially result in an increased haemolytic activity84, this does not appear to apply in this case, which may be explained by the relatively flexible non-helical N-terminal and C-terminal segments, which combined account for 10 of the sequences 31 residues. This flexibility can be seen in the root-mean-square fluctuation (RMSF) plot, where the peptide’s terminal residues have greater RMSF values than the residues located within the \(\alpha\)-helical segment (Fig. 8).
Conclusion
A number of biologically active peptides have been isolated from the American bullfrog Lithobates catesbeianus. The isolated peptides are diverse, and not limited to a single peptide family; palustrin-Ca is the only palustrin peptide isolated from this amphibian. Palustrin-Ca can be considered to be a member of the palustrin-2 family of peptides, sharing an average 42.09% sequence identity with 18 peptides of the palustrin-2 family. Peptides that belong to the palustrin-2 family exhibit a variety of potent biological activities, including antimicrobial and anticancer activities, and therefore represent exciting candidates for novel drug development.
Palustrin-Ca is 31 amino acid residues long, of which 5 are lysine, 4 are glycine and 4 are leucine. The peptide has a cyclic disulfide-bridged heptapeptide domain at its C-terminus, which is conserved with other peptides of the palustrin-2 family.
This study employed NMR spectroscopy and molecular modelling methods to characterise the peptide’s three-dimensional structure and obtain an ensemble of model structures. To date, palustrin-Ca is the only palustrin peptide to have had its three-dimensional structure elucidated. The results established that palustrin-Ca possesses an amphipathic, \(\alpha\)-helical structure between residues Ile\(^{6}\)-Ala\(^{26}\) in a 50% TFE-H\(_{2}\)O mixed solvent system, with the terminal segments predominantly defined as turns, with significant coil character as well.
A molecular dynamics simulation was conducted, whereby the medoid NMR-determined structure was simulated with an SDS micelle. The simulation results suggest that palustrin-Ca preferentially adopts a position parallel to the micelle’s surface. This configuration is most energetically favourable as the peptide’s hydrophobic residues can penetrate to the micelle’s hydrophobic core, while the hydrophilic residues remain in contact with the aqueous solvent. The RDF plot in Fig. 7 illustrates this clearly; the greatest values for g(r) at small distances are observed for the hydrophobic Leu, Ile, Ala and Phe residues, demonstrating the importance of hydrophobicity for a peptide’s ability to bind the target membrane. The observed preference for a position parallel to the micelle surface indicates that the peptide most probably exerts its antimicrobial activity through a non-pore-forming mechanism of action, such as the carpet model or the interfacial activity model.
This study’s results will facilitate future work focused on investigating HDP structures, and their interactions with zwitterionic lipid bilayers, in order to further our understanding of the relationships between HDP structure and function. This study has extended earlier work from our research group that led to the elucidation of a number of host-defence peptide structures, and also complements related bioinformatic studies on peptide structure and function85,86,87,88.
References
Xia, X., Cheng, L., Zhang, S., Wang, L. & Hu, J. The role of natural antimicrobial peptides during infection and chronic inflammation. Antonie Van Leeuwenhoek 111, 5–26 (2018).
Maróti Gergely, G., Kereszt, A., Kondorosi, É. & Mergaert, P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 162, 363–374 (2011).
Avila, E. E. Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 18, 1098–1119 (2017).
da Silva Pereira, L. et al. Characterization of Capsicum annuum L. leaf and root antimicrobial peptides: Antimicrobial activity against phytopathogenic microorganisms. Acta Physiologiae Plantarum 40, 107 (2018).
El Samak, M., Solyman, S. M. & Hanora, A. Antimicrobial activity of bacteria isolated from Red Sea marine invertebrates. Biotechnol. Rep. 19, e00275 (2018).
Gordon, Y. J., Romanowski, E. G. & McDermott, A. M. Mini review: A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 30, 505–515 (2005).
Tanphaichitr, N. et al. Potential use of antimicrobial peptides as vaginal spermicides/microbicides. Pharmaceuticals 9, 13 (2016).
Gaspar, D., Salomé Veiga, A. & Castanho, M. A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 4, 294 (2013).
Agarwal, G. & Gabrani, R. Antiviral Peptides: Identification and Validation. Int. J. Pept. Res. Ther. 27, 149–168 (2021).
Lacerda, A. F., Pelegrini, P. B., De Oliveira, D. M., Vasconcelos, É. A. & Grossi-de Sá, M. F. Anti-parasitic peptides from arthropods and their application in drug therapy. Front. Microbiol. 7, 91 (2016).
Zhao, R.-L., Han, J.-Y., Han, W.-Y., He, H.-X. & Ma, J.-F. Effects of two novel peptides from skin of lithobates catesbeianus on tumor cell morphology and proliferation. In Molecular Cloning—Selected Applications in Medicine and Biology (InTech, 2011). http://aps.unmc.edu/AP.
Lai, R., Liu, H., Hui Lee, W. & Zhang, Y. An anionic antimicrobial peptide from toad Bombina maxima. Biochem. Biophys. Res. Commun. 295, 796–799 (2002).
Zelezetsky, I. & Tossi, A. Alpha-helical antimicrobial peptides—Using a sequence template to guide structure–activity relationship studies. Biochim. Biophys. Acta Biomembr. 1758, 1436–1449 (2006).
Xie, J. et al. Novel antimicrobial peptide CPF-C1 analogs with superior stabilities and activities against multidrug-resistant bacteria. Chem. Biol. Drug Des. 90, 690–702 (2017).
Yin, L. M., Edwards, M. A., Li, J., Yip, C. M. & Deber, C. M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 287, 7738–7745 (2012).
Son, M., Lee, Y., Hwang, H., Hyun, S. & Yu, J. Disruption of interactions between hydrophobic residues on nonpolar faces is a key determinant in decreasing hemolysis and increasing antimicrobial activities of \(\alpha\)-helical amphipathic peptides. ChemMedChem 8, 1638–1642 (2013).
Hornef, M. W., Pütsep, K., Karlsson, J., Refai, E. & Andersson, M. Increased diversity of intestinal antimicrobial peptides by covalent dimer formation. Nat. Immunol. 5, 836–843 (2004).
Lorenzon, E. N., Piccoli, J. P., Santos-Filho, N. A. & Cilli, E. M. Dimerization of antimicrobial peptides: A promising strategy to enhance antimicrobial peptide activity. Protein Pept. Lett. 26, 98–107 (2019).
Beveridge, T. J. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 (1999).
Weidenmaier, C. & Peschel, A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6, 276–287 (2008).
Zanin, L. M. P. et al. Interaction of a synthetic antimicrobial peptide with model membrane by fluorescence spectroscopy. Eur. Biophys. J. 42, 819–831 (2013).
Hong, S. Y., Park, T. G. & Lee, K. H. The effect of charge increase on the specificity and activity of a short antimicrobial peptide. Peptides 22, 1669–1674 (2001).
Yeaman, M. R. & Yount, N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 (2003).
Perron, G. G., Zasloff, M. & Bell, G. Experimental evolution of resistance to an antimicrobial peptide. Proc. R. Soc. B Biol. Sci. 273, 251–256 (2006).
Hall, K., Lee, T. H., Mechler, A. I., Swann, M. J. & Aguilar, M. I. Real-time measurement of membrane conformational states induced by antimicrobial peptides: Balance between recovery and lysis. Sci. Rep. 4, 1–9 (2014).
Sani, M. A., Henriques, S. T., Weber, D. & Separovic, F. Bacteria may cope differently from similar membrane damage caused by the Australian tree frog antimicrobial peptide maculatin 1.1. J. Biol. Chem. 290, 19853–19862 (2015).
Brogden, K. A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria?. Nat. Rev. Microbiol. 3, 238–250 (2005).
Shai, Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. Orig. Res. Biomol. 66(4), 236–248 (2002).
Hammond, K. et al. Switching cytolytic nanopores into antimicrobial fractal ruptures by a single side chain mutation. ACS Nano 15, 9679–9689. https://doi.org/10.1021/acsnano.1c00218 (2021).
Sani, M. A. & Separovic, F. How membrane-active peptides get into lipid membranes. Acc. Chem. Res. 49, 1130–1138. https://doi.org/10.1021/acs.accounts.6b00074 (2016).
Subasinghage, A. P., Conlon, J. M. & Hewage, C. M. Conformational analysis of the broad-spectrum antibacterial peptide, ranatuerin-2CSa: Identification of a full length helix-turn-helix motif. Biochim. Biophys. Acta Proteins Proteomics 1784, 924–929 (2008).
Subasinghage, A. P., Conlon, J. M. & Hewage, C. M. Development of potent anti-infective agents from Silurana tropicalis: Conformational analysis of the amphipathic, alpha-helical antimicrobial peptide XT-7 and its non-haemolytic analogue [G4K]XT-7. Biochim. Biophys. Acta Proteins Proteomics 1804, 1020–1028. https://doi.org/10.1016/j.bbapap.2010.01.015 (2010).
Subasinghage, A. P., O’Flynn, D., Conlon, J. M. & Hewage, C. M. Conformational and membrane interaction studies of the antimicrobial peptide alyteserin-1c and its analogue [E4K]alyteserin-1c. Biochim. Biophys. Acta Biomembr. 1808, 1975–1984. https://doi.org/10.1016/j.bbamem.2011.04.012 (2011).
Timmons, P. B., O’Flynn, D., Conlon, J. M. & Hewage, C. M. Structural and positional studies of the antimicrobial peptide brevinin-1BYa in membrane-mimetic environments. J. Pept. Sci. 25, e3208 (2019).
Timmons, P. B., O’Flynn, D., Conlon, J. M. & Hewage, C. M. Insights into conformation and membrane interactions of the acyclic and dicarba-bridged brevinin-1BYa antimicrobial peptides. Eur. Biophys. J. 48, 701–710. https://doi.org/10.1007/s00249-019-01395-y (2019).
Benetti, S., Timmons, P. B. & Hewage, C. M. NMR model structure of the antimicrobial peptide maximin 3. Eur. Biophys. J. 48, 203–212. https://doi.org/10.1007/s00249-019-01346-7 (2019).
Timmons, P. B. & Hewage, C. M. Biophysical study of the structure and dynamics of the antimicrobial peptide maximin 1. J. Pept. Sci. 27, e3370 (2021).
Bax, A. & Davis, D. G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Resonance (1969) 65, 355–360 (1985).
Kumar, A., Ernst, R. R. & Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Top. Catal. 95, 1–6 (1980).
John, B. K., Plant, D., Webb, P. & Hurd, R. E. Effective combination of gradients and crafted RF pulses for water suppression in biological samples. J. Magn. Resonance 1969(98), 200–206 (1992).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Güntert, P., Braun, W. & Wüthrich, K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 217, 517–530 (1991).
Güntert, P., Mumenthaler, C. & Wüthrich, K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 (1997).
Güntert, P. & Buchner, L. Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR 62, 453–471. https://doi.org/10.1007/s10858-015-9924-9 (2015).
Mackerell, A. D., Feig, M. & Brooks, C. L. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulation. J. Comput. Chem. 25, 1400–1415 (2004).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802. https://doi.org/10.1002/jcc.20289 (2005).
Humphrey, W., Dalke, A. & Schulten, K. V. M. D. Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Berman, H., Henrick, K. & Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 10, 980 (2003).
Jakobtorweihen, S., Ingram, T. & Smirnova, I. Combination of COSMOmic and molecular dynamics simulations for the calculation of membrane-water partition coefficients. J. Comput. Chem. 34, 1332–1340. https://doi.org/10.1002/jcc.23262 (2013).
MacKerell, A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616. https://doi.org/10.1021/jp973084f (1998).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189. https://doi.org/10.1063/1.467468 (1994).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 103, 4613–4621. https://doi.org/10.1063/1.470648 (1995).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N\(\cdot\)log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092. https://doi.org/10.1063/1.464397 (1993).
Ryckaert, J. P., Ciccotti, G. & Berendsen, H. J. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Sönnichsen, F. D., Van Eyk, J. E., Hodges, R. S. & Sykes, B. D. Effect of trifluoroethanol on protein secondary structure: An NMR and CD study using a synthetic actin peptide. Biochemistry 31, 8790–8798 (1992).
Goodman, M. & Rosen, I. G. Conformational aspects of polypeptide structure XVI. Rotatory constants, cotton effects, and ultraviolet absorption data for glutamate oligomers and co-oligomers. Biopolymers 2, 537–559 (1964).
Buck, M. Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31, 297–355 (1998).
Bodkin, M. J. & Goodfellow, J. M. Hydrophobic solvation in aqueous trifluoroethanol solution. Biopolymers 39, 43–50 (1996).
Yi, G. S., Park, C. B., Kim, S. C. & Cheong, C. Solution structure of an antimicrobial peptide buforin II. FEBS Letters 398, 87–90 (1996).
Seo, M. D., Won, H. S., Kim, J. H., Mishig-Ochir, T. & Lee, B. J. Antimicrobial peptides for therapeutic applications: A review. Molecules 17, 12276–12286 (2012).
Raj, P. A., Marcus, E. & Sukumaran, D. K. Structure of human salivary histatin 5 in aqueous and nonaqueous solutions. Biopolymers 45, 51–67 (1998).
Gong, Z., Ikonomova, S. P. & Karlsson, A. J. Secondary structure of cell-penetrating peptides during interaction with fungal cells. Protein Sci. 27, 702–713. https://doi.org/10.1002/pro.3364 (2018).
Pirtskhalava, M., Vishnepolsky, B., Grigolava, M. & Managadze, G. Physicochemical features and peculiarities of interaction of amp with the membrane. Pharmaceuticals 14, 471 (2021).
Shiraki, K., Nishikawa, K. & Goto, Y. Trifluoroethanol-induced stabilization of the \(\alpha\)-helical structure of \(\beta\)-lactoglobulin: Implication for non-hierarchical protein folding. J. Mol. Biol. 245, 180–194 (1995).
Venneti, K. C. & Hewage, C. M. Conformational and molecular interaction studies of glucagon-like peptide-2 with its N-terminal extracellular receptor domain. FEBS Lett. 585, 346–352 (2011).
Rajan, R. & Balaram, P. A model for the interaction of trifluoroethanol with peptides and proteins. Int. J. Pept. Protein Res. 48, 328–336 (1996).
Roccatano, D., Colombo, G., Fioroni, M. & Mark, A. E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: A molecular dynamics study. Proc. Natl. Acad. Sci. USA 99, 12179–12184 (2002).
Marion, D., Zasloff, M. & Bax, A. A two-dimensional NMR study of the antimicrobial peptide magainin 2. FEBS Lett. 227, 21–26 (1988).
Toke, O. et al. A kinked antimicrobial peptide from Bombina maxima. I. Three-dimensional structure determined by NMR in membrane-mimicking environments. Eur. Biophys. J. 40, 447–462 (2011).
Alaña, I., Malthouse, J. P. G., O’Harte, F. P. & Hewage, C. M. The bioactive conformation of glucose-dependent insulinotropic polypeptide by NMR and CD spectroscopy. Proteins Struct. Funct. Genet. 68, 92–99. https://doi.org/10.1002/prot.21372 (2007).
Wagner, G., Pardi, A. & Wüthrich, K. Hydrogen bond length and 1H NMR chemical shifts in proteins. J. Am. Chem. Soc. 105, 5948–5949 (1983).
Wishart, D. S., Sykes, B. D. & Richards, F. M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31, 1647–1651 (1992).
Frishman, D. & Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Bioinform. 23, 566–579 (1995).
Tossi, A., Sandri, L. & Giangaspero, A. Amphipathic, \(\alpha\)-helical antimicrobial peptides. Biopolymers 55, 4–30 https://doi.org/10.1002/1097-0282(2000)55:1%3c4::AID-BIP30%3e3.0.CO;2-M (2000).
Langham, A. & Kaznessis, Y. N. Molecular simulations of antimicrobial peptides. Methods Mol. Biol. 618, 267–285 (2010).
Khandelia, H., Langham, A. A. & Kaznessis, Y. N. Driving engineering of novel antimicrobial peptides from simulations of peptide-micelle interactions. Biochim. Biophys. Acta Biomembr. 1758, 1224–1234 (2006).
Clark, T. D., Bartolotti, L. & Hicks, R. P. The application of DOSY NMR and molecular dynamics simulations to explore the mechanism(s) of micelle binding of antimicrobial peptides containing unnatural amino acids. Biopolymers 99, 548–561 (2013).
Roussel, G. et al. Peptide-surfactant interactions: A combined spectroscopic and molecular dynamics simulation approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 190, 464–470 (2018).
Crusca, E. et al. NMR structures and molecular dynamics simulation of hylin-a1 peptide analogs interacting with micelles. J. Pept. Sci. 23, 421–430 (2017).
Dathe, M. & Wieprecht, T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta Biomembr. 1462, 71–87 (1999).
Shai, Y. & Oren, Z. From, “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides 22, 1629–1641 (2001).
Chen, Y. et al. Role of peptide hydrophobicity in the mechanism of action of \(\alpha\)-helical antimicrobial peptides. Antimicrob. Agents Chemother. 51, 1398–1406 (2007).
Idiong, G. et al. Investigating the effect of a single glycine to alanine substitution on interactions of antimicrobial peptide latarcin 2a with a lipid membrane. Eur. Biophys. J 40, 1087–1100. https://doi.org/10.1007/s00249-011-0726-z (2011).
Timmons, P. B. & Hewage, C. M. HAPPENN is a novel tool for hemolytic activity prediction for therapeutic peptides which employs neural networks. Sci. Rep. 10, 10869 (2020).
Timmons, P. B. & Hewage, C. M. ENNAACT is a novel tool which employs neural networks for anticancer activity classification for therapeutic peptides. Biomed. Pharmacother. 133, 111051 https://doi.org/10.1016/j.biopha.2020.111051 (2021).
Timmons, P. B. & Hewage, C. M. ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides. Brief. Bioinform. 22, bbab258. https://doi.org/10.1093/bib/bbab258 (2021).
Timmons, P. B. & Hewage, C. M. APPTEST is a novel protocol for the automatic prediction of peptide tertiary structures. Brief. Bioinform. 22, bbab308. https://doi.org/10.1093/bib/bbab308 (2021).
Acknowledgements
CH is grateful to John O’Brien and Manuel Ruether at Trinity College Dublin for NMR facilities, University College Dublin for a Research Scholarship to PBT, ICHEC for access to supercomputer facilities. The solution structure of palustrin-Ca was deposited to the PDB at the RSCB with deposition code 7P4X.
Author information
Authors and Affiliations
Contributions
P.B.T. and C.M.H. determined the scope of the experiments and wrote the manuscript. P.B.T. analysed the spectra, generated the model structure and performed the molecular dynamics simulations. Both authors contributed to and have approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Timmons, P.B., Hewage, C.M. Conformation and membrane interaction studies of the potent antimicrobial and anticancer peptide palustrin-Ca. Sci Rep 11, 22468 (2021). https://doi.org/10.1038/s41598-021-01769-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01769-3
This article is cited by
-
Recent advances and applications of peptide–agent conjugates for targeting tumor cells
Journal of Cancer Research and Clinical Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.