Abstract
Recent studies have shown that adipose tissue is an immunological organ. While inflammation in energy-storing white adipose tissues has been the focus of intense research, the regulatory mechanisms of inflammation in heat-producing brown adipose tissues remain largely unknown. We previously identified apoptosis signal-regulating kinase 1 (ASK1) as a critical regulator of brown adipocyte maturation; the PKA-ASK1-p38 axis facilitates uncoupling protein 1 (UCP1) induction cell-autonomously. Here, we show that ASK1 suppresses an innate immune pathway and contributes to maintenance of brown adipocytes. We report a novel chemical pull-down method for endogenous kinases using analog sensitive kinase allele (ASKA) technology and identify an ASK1 interactor in brown adipocytes, receptor-interacting serine/threonine-protein kinase 2 (RIPK2). ASK1 disrupts the RIPK2 signaling complex and inhibits the NOD-RIPK2 pathway to downregulate the production of inflammatory cytokines. As a potential biological significance, an in vitro model for intercellular regulation suggests that ASK1 facilitates the expression of UCP1 through the suppression of inflammatory cytokine production. In parallel to our previous report on the PKA-ASK1-p38 axis, our work raises the possibility of an auxiliary role of ASK1 in brown adipocyte maintenance through neutralizing the thermogenesis-suppressive effect of the NOD-RIPK2 pathway.
Similar content being viewed by others
Introduction
Growing evidence suggests that adipose tissue is an immunological organ. While adipose tissue has long been merely regarded as a lipid-storing organ, it is now widely recognized that adipose tissue expresses various receptors for cytokines and chemokines and responds to proinflammatory mediators secreted by itself1,2. Physiologically, low-grade chronic inflammation is observed under obesity and is strongly implicated in the onset and development of obesity-related diseases such as type 2 diabetes and cardiovascular disease3. Therefore, controlling inflammatory signaling in adipose tissue would be a potential target to combat obesity and obesity-induced diseases.
Adipose tissues in mammals can be classified into two types: white adipose tissue (WAT) and brown adipose tissue (BAT). While the major function of white adipocytes is to store excess energy as triglycerides, brown adipocytes uniquely express uncoupling protein 1 (UCP1), which is a principal contributor to its unique function in nonshivering thermogenesis4,5. BAT is less susceptible to inflammation than WAT, but sustained overnutrition ultimately induces a proinflammatory environment in BAT and results in impaired thermogenic machinery of brown adipocytes6. BAT from diet-induced obese mice showed increased infiltration of immune cells, as well as upregulation of proinflammatory cytokines7. Cold-induced UCP1 induction was suppressed in adipose tissue from obese mice8. Hence, these recent studies suggest that obesity-induced inflammation leads to dysfunction of brown adipocytes through the reduction of UCP1 and other thermogenic markers. However, the regulatory mechanisms of inflammation in brown adipocytes remain largely obscure.
The NOD-RIPK2 pathway plays a crucial role in host defense against bacterial infection and is associated with the onset of autoimmune disorders9. In a cell under bacterial infection, intracellular pattern recognition receptors sense the peptidoglycan derivatives of bacterial cell wall; that is, nucleotide-binding oligomerization domain 1 (NOD1) and NOD2 recognize meso-diaminopimelic acid (DAP) and muramyl dipeptide (MDP), respectively. Upon ligand binding, NODs oligomerize through the caspase recruitment domain (CARD) and induce further oligomerization of another CARD-containing protein, receptor-interacting serine/threonine-protein kinase 2 (RIPK2). Oligomerized RIPK2 is K63-polyubiquitinated by X-linked inhibitor of apoptosis protein (XIAP), linear ubiquitin chain assembly complex (LUBAC), and other E3 ligases and further recruits its downstream effectors, including TGF-beta activated kinase 1 (TAK1)/TAK1 binding protein (TAB) complex and nuclear factor of kappa B (NF-κB) essential modulator (NEMO) complex. Consequently, the c-jun N-terminal kinase (JNK), p38 mitogen-activated protein kinase (MAPK) and NF-κB pathways are activated, leading to the induction of proinflammatory cytokines10.
In addition to the role in immune cells, the NOD-RIPK2 pathway is implicated in adipose inflammation and affects the physiology of adipocytes. In adipocytes, pattern recognition receptors including NOD1 are considered to be activated by bacterial fragments translocated from gut microbiota11, which is augmented under obesity12. NOD1 activation in white adipocytes induces insulin resistance and lipolysis13,14,15 and suppresses adipocyte differentiation with attenuated expression of adipocyte markers and lipid accumulation16. Moreover, NOD1 activation in brown adipocytes leads to suppression of brown adipocyte markers, including UCP117. These lines of evidence suggest that the inflammatory NOD-RIPK2 pathway in adipocytes suppresses the differentiation of adipocytes.
We have previously reported apoptosis signal-regulating kinase 1 (ASK1)18 as a critical regulator of thermogenesis; under β-adrenergic receptor stimulation, protein kinase A (PKA) activates the ASK1-p38 MAPK axis to induce brown adipocyte-specific genes19,20. Here, we show that ASK1 suppresses the NOD-RIPK2 pathway in brown adipocytes. We report an analog sensitive kinase allele (ASKA) technology-based pull-down mass spectrometry (MS) method and identify RIPK2 as a novel interactor of ASK1 in brown adipocytes. ASK1 interferes with the NOD-RIPK2 pathway by inhibiting the activation of the RIPK2 signaling complex. As a potential biological significance, our in vitro model for intercellular thermogenic regulation implies that the suppressive function of ASK1 in the NOD-RIPK2 pathway positively contributes to the maintenance of thermogenic function in BAT under inflammation, which suggests a complementary role to the ASK1’s function as a positive regulator of BAT thermogenesis via PKA-ASK1-p38 axis. This work demonstrates an example application of our novel chemical pull-down method and reveals the multifaceted finetuning role of ASK1 in brown adipocytes.
Results
ASKA technology-based pull-down MS method identified RIPK2 as an interactor of ASK1
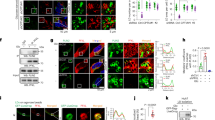
ASK1 forms a mega-Dalton complex (ASK1 signalosome) in a cell21. To explore unrevealed mechanisms or functions of ASK1 in BAT, we first sought to identify components of the ASK1 signalosome in brown adipocytes. ASKA technology is a kinase modification method that was originally developed to specifically inhibit a genetically modified kinase (as-kinase) with the ATP analog 1NA-PP122; while bulky 1NA-PP1 cannot enter the ATP-binding pocket of wild-type kinases, the modification to substitute less bulky amino acids for residues in the hydrophobic gatekeeper region of ATP-binding pocket enables 1NA-PP1 to enter the ATP-binding pocket of as-kinase and to compete with ATP for the as-kinase. We have previously generated Ask1ASKA knock-in mice harboring an ASKA of Ask1 and demonstrated that primary cells from Ask1ASKA knock-in mice showed expression and activation levels of ASK1 comparable to those from wild-type mice23. In this study, by leveraging the highly specific binding affinity of 1NA-PP1 to the as-kinase, we developed a chemical pull-down assay for an endogenous kinase, referred to as the “ASKA pull-down MS method” (Fig. 1a). In brief, the endogenous as-kinase signalosome was pulled down by incubating tissue/cell extracts from ASKA knock-in mice with 1NA-PP1-bound carrier beads, eluted by adding excess free 1NA-PP1, and subjected to MS analysis. To estimate the optimal linker length between 1NA-PP1 and its carrier bead, we checked the ATP-binding pocket of the ASK1 kinase domain by analyzing the previously reported crystal structure24 (Fig. 1b). Based on the assumed depth of the ATP-binding pocket, we synthesized two 1NA-PP1 derivatives with different linker lengths (1NA-PP1-Lx, x ∈ {1, 2}, Fig. 1c, Supplementary Note). Of note, the carrier beads we used have an approximately 20 Å linker with the N-hydroxysuccinimide reactive group, which cross-links with each 1NA-PP1-Lx. Using a surface plasmon resonance (SPR) assay, we confirmed the direct biophysical affinity of 1NA-PP1-Lx with the recombinant as-ASK1 kinase domain (KD) in vitro but not with wild-type ASK1 KD (Fig. 1d), validating that our pull-down strategy specifically captures as-kinase. In addition, because the analyte ASK1 KD can be dimerized in solution24, we modeled the bivalent analyte model, which fit our SPR data well. The dissociation constant for the first phase (KD1) of 1NA-PP1-L1 or 1NA-PP1-L2 vs. as-ASK1 KD was calculated as KD1 = 2.06 × 10−6 [M] or 2.23 × 10−6 [M], respectively, implying that this affinity is within a suitable range not only for pull-down but also for the subsequent elution step (Fig. 1a). We next compared the pull-down capacity of each 1NA-PP1 derivative for as-ASK1 in tissue lysates derived from Ask1ASKA knock-in mice. Interestingly, while 1NA-PP1-L2-immobilized beads successfully pulled down as-ASK1 from brain samples, 1NA-PP1-L1-immobilized beads failed to capture as-ASK1 (Fig. 1e). This discrepancy between the direct biophysical affinity and the pull-down capacity of 1NA-PP1-L1 probably stems from the accessibility of 1NA-PP1-L1 to the ATP-binding pocket; since the entry of the 1NA-PP1 group to the ATP-binding pocket of as-ASK1 may be hindered by physical repulsion of the carrier bead with the components of the signalosome or with C/N-terminal regions of ASK1, 1NA-PP1-L1 would be too short to capture as-ASK1 in the signalosome. Hence, we adopted 1NA-PP1-L2 for the following pull-down experiments. In the final step of the purification procedures, as-ASK1 was effectively eluted from the incubated 1NA-PP1-L2-immobilized carrier beads by competitive elution with free 1NA-PP1 (Fig. 1f). To examine whether as-ASK1 maintains an intact signalosome after the series of purification procedures, we performed a size exclusion chromatography analysis and compared the as-ASK1 signalosomes before and after purification. After purification from primary brown adipocytes of Ask1ASKA knock-in mice, as-ASK1 was observed in the same fractions of as-ASK1 before purification (Fig. 1g), suggesting that the as-ASK1 signalosome is kept intact throughout all purification steps. Therefore, as-ASK1 signalosomes purified from primary brown adipocytes of Ask1ASKA knock-in mice were subjected to MS analysis. Comparing the MS results of 1NA-PP1-eluted samples with that of the DMSO-eluted negative control, 32 candidates were identified as interactors of ASK1 in brown adipocytes (Table 1). Among them, previously reported ASK1 interactors were included, such as ASK2 (also known as MAP3K6), a member of the ASK family25, and 14-3-3γ (also known as YWHAG), a negative regulator of ASK126. In contrast to the ASK1 interactor candidates identified by the Flag-tag pull-down MS of Flag-tagged ASK1-overexpressing HEK293 cells in previous reports27,28, the identified ASK1 interactor candidates were relatively unique, with 30 out of 32 candidates categorized as brown adipocyte-specific interactors (Fig. 1h, Table 1), implying that our novel method listed up brown-specific ASK1 interactor candidates. We further validated the interaction between ASK1 and an ASK1 interactor candidate with coimmunoprecipitation assay. By immunoprecipitating RIPK2, one of the brown-specific interactor candidates, the coimmunoprecipitation of endogenous ASK1 was confirmed in the brown adipose cell line HIB 1B29 (Fig. 1i), supporting the validation of our ASKA pull-down MS method. Of note, it might have been possible to regard RIPK2 as an artifact in our system because RIPK2 was reported to bind directly to 1NA-PP130, but this data using other method suggests that RIPK2 is a true positive interactor of ASK1.
The ASKA technology-based pull-down MS method identified RIPK2 as an interactor of ASK1. (a) Overview of the ASKA pull-down MS method. The endogenous as-kinase signalosome was pulled down by incubating tissue/cell extracts from ASKA knock-in mice with 1NA-PP1-bound carrier beads, eluted by adding excess free 1NA-PP1, and subjected to mass spectrometry analysis. X and Y indicate unidentified components of the signalosome. (b) ATP-binding pocket of the ASK1 kinase domain (PDB: 2CLQ)24. (c) Chemical structures of 1NA-PP1 derivatives (1NA-PP1-Lx, x ∈ {1, 2}). 1NA-PP1 a bulky analog of ATP. (d) Binding affinity between the as-ASK1 kinase domain (KD) and 1NA-PP1-Lx. Setting 1NA-PP1-Lx as the ligand, the surface plasmon resonance signal was measured under a series of concentrations of the ASK1 KD analytes (white rectangle period: buffer, black rectangle period C1: 0.04 µM, C2: 0.06 µM, C3: 0.09 µM, C4: 0.13 µM, C5: 0.20 µM). R.U. resonance unit. (e) Pull-down ability of 1NA-PP1-Lx-immobilized beads for endogenous as-ASK1 in tissue lysate. Fractionated brain extracts from Ask1ASKA knock-in mice were incubated with 1NA-PP1-Lx-immobilized beads, and the incubated beads and flow-through samples were subjected to immunoblotting (IB). Note that superfluous lanes were digitally eliminated from blot images as indicated by black lines and the uncropped images are presented in Supplementary Data. (f) Specific elution of as-ASK1 with 1NA-PP1. as-ASK1 was pulled down from the cell lysate of Ask1ASKA knock-in mouse-derived primary brown adipocytes using 1NA-PP1-L2-immobilized beads and eluted with excess free 1NA-PP1. DMSO solvent of 1NA-PP1, a negative control. (g) Size of the as-ASK1 signalosome before and after purification. Cell lysates from primary brown adipocytes of Ask1ASKA knock-in mice before and after purification with 1NA-PP1-L2-immobilized beads and 1NA-PP1 were fractionated with size exclusion chromatography. (h) Venn diagram of the ASK1 interactor candidates. ASKA ASKA pull-down MS for as-ASK1 derived from primary brown adipocytes in Ask1ASKA knock-in mice, FLAG-1 Flag-tag pull-down MS for Flag-ASK1 derived from overexpressed HEK293A cells27, FLAG-2 Flag-tag pull-down MS for ASK1-HA-Flag derived from overexpressed HEK293 cells28. See Table 1 for each protein list. (i) Interaction between RIPK2 and ASK1 in a brown adipose cell line. Differentiated HIB 1B cells were processed for immunoprecipitation with anti-RIPK2 antibody. IP immunoprecipitation. *Immunoglobulin G heavy chain.
ASK1 inhibits the activation of the RIPK2 signaling complex
Among the ASK1 interactor candidates, we specifically focused on RIPK2, a critical adaptor molecule in the inflammatory NOD-RIPK2 pathway because it is implicated in adipose inflammation in brown adipocytes17 and the interaction between ASK1 and RIPK2 in brown adipocytes suggests a potential involvement of ASK1 in brown adipose inflammation. We first selected HEK293A cells as an experimental model to investigate the functional relationship between ASK1 and the NOD-RIPK2 pathway and its molecular mechanism because HEK293A cells enable us to utilize overexpression approach with high efficiency. Upon ligand binding to NOD receptors, RIPK2 recruits its effectors, including XIAP and the TAB/TAK1 complex, and activates downstream MAPK and NF-κB signaling to induce proinflammatory responses31. Due to the low expression level of endogenous NOD1, stimulation with a synthetic NOD1 ligand C12-iE-DAP32 failed to activate JNK and p38 MAPK in the original HEK293A cells (data not shown; cf. lanes 1 and 2 in Fig. 2a). Therefore, we established a stable transfectant HEK293A cell line that expresses Myc-tagged mouse NOD1 under tetracycline treatment (NOD1-HEK293A cells). C12-iE-DAP stimulation activated the NOD-RIPK2 pathway, as indicated by enhanced phosphorylation levels of JNK and p38 MAPK in tetracycline-treated NOD1-HEK293A cells (Fig. 2a). The interaction between ASK1 and RIPK2 was confirmed in the NOD1-HEK293A cell line (Fig. 2b). Then, we investigated whether RIPK2 acts on ASK1 or vice versa. In contrast to an oxidative stress inducer H2O2, a potent ASK1 activator33, C12-iE-DAP did not induce ASK1 phosphorylation in NOD1-HEK293A cells (Fig. 2a), suggesting that activation of the NOD-RIPK2 pathway does not affect the kinase activity of ASK1. In contrast, overexpression of ASK1 suppressed the degradation of NF-κB inhibitor, alpha (IκBα) under C12-iE-DAP stimulation (Fig. 2c). Note that the activation of the NOD-RIPK2 pathway was monitored with the degradation of IκBα here because exogenously expressed ASK1 drastically increases the phosphorylation level of JNK and p38 MAPK34. Moreover, the knockdown of ASK1 enhanced C12-iE-DAP-induced IκBα degradation in NOD1-HEK293A cells (Fig. 2d). These results suggested that ASK1 suppresses the activation of the NOD-RIPK2 pathway.
ASK1 inhibits the activation of the RIPK2 signaling complex. (a) Phosphorylation levels of ASK1 under NOD1 ligand treatment in NOD1-6Myc-stably expressing HEK293A cells (NOD1-HEK293A cells). NOD1-HEK293A cells were stimulated with a NOD1 ligand C12-iE-DAP (1 µg/mL, 30 min). mNOD1 mouse NOD1, H2O2 a positive control of ASK1 phosphorylation, 250 µM, 30 min. *Nonspecific bands. (b) Interaction between RIPK2 and ASK1 in NOD1-HEK293A cells. After the induction of NOD1 with tetracycline, the co-transfected NOD1-HEK293A cells were processed for immunoprecipitation by anti-Flag-tag antibody beads. mASK1 mouse ASK1, mRIPK2 mouse RIPK2. (c,d) Effects of ASK1 overexpression (c) or ASK1 knockdown (d) on NOD-RIPK2 pathway activation. ASK1-overexpressing (c) or ASK1-knockdown (d) NOD1-HEK293A cells were stimulated with C12-iE-DAP (1 µg/mL) for 60 min or the indicated periods of time, respectively. Note that cycloheximide (50 µg/mL) was treated to prevent the rapid feedback synthesis of IκBα. hASK1 human ASK1. (e) Schematic representation of RIPK2 deletion mutants. The numbers indicate the amino acid (a.a.) positions in wild-type (WT). Black rectangles indicate registered domains in the Pfam database (P58801). KD kinase domain, IM intermediate domain, CARD caspase-recruitment domain. (f) Binding ability of RIPK2 domain mutants to ASK1. Co-transfected HEK293A cells were processed for immunoprecipitation by anti-Flag-tag antibody beads. (g) Effects of ASK1 overexpression on the RIPK2-XIAP interaction. Co-transfected HEK293A cells were processed for immunoprecipitation by anti-Flag-tag antibody beads. mXIAP mouse XIAP. *Immunoglobulin G heavy chain. (h) Effects of ASK1 knockdown on K63-polyubiquitination of endogenous RIPK2 under NOD1 ligand stimulation. ASK1-knockdown NOD1-HEK293A cells were stimulated with C12-iE-DAP (1 µg/mL) for the indicated times, from which lysate K63-polyubiquitinated conjugates were purified with a tandem ubiquitin binding entity (TUBE) pull-down assay. Note that superfluous lanes were digitally eliminated from blot images in (c,g,h) as indicated by black lines and the uncropped images are presented in Supplementary Data.
To explore the inhibitory mechanism of the NOD-RIPK2 pathway by ASK1, we determined the ASK1-interacting domain of RIPK2 by coimmunoprecipitation analysis. RIPK2 is composed of three domains: the kinase domain (KD), intermediate domain (IM), and caspase recruitment domain (CARD)35 (Fig. 2e). Coimmunoprecipitation of wild-type ASK1 with RIPK2 domain mutants revealed that ASK1 specifically bound to KD of RIPK2 (Fig. 2f), which contains an essential residue for K63-polyubiquitination and subsequent recruitment of the TAB/TAK1 and IκB kinase (IKK) complexes36. Hence, we hypothesized that ASK1 inhibits the NOD-RIPK2 pathway by physically interfering with the formation of the RIPK2 complex. As an interfering effect of ASK1 on the RIPK2 complex, we evaluated the interaction between RIPK2 and one of its E3-ligases XIAP, a major contributor to K63-polyubiquitination on RIPK2 upon pathway activation37. The amount of XIAP that coimmunoprecipitated with RIPK2 was reduced by co-overexpression of ASK1 (Fig. 2g), suggesting that ASK1 competes with XIAP for RIPK2 interaction. By using a tandem ubiquitin binding entity (TUBE) pull-down assay38, we assessed the K63-polyubiquitination of RIPK2, which is induced in an early step of RIPK2 signaling complex formation upon NOD1 ligand stimulation36. Knockdown of ASK1 enhanced the K63-polyubiquitination of endogenous RIPK2 upon C12-iE-DAP stimulation in NOD1-HEK293A cells (Fig. 2h). These data collectively suggest that ASK1 downregulates the NOD-RIPK2 pathway at least in part by impeding RIPK2 from forming a functional signaling complex.
ASK1 suppresses the NOD-RIPK2 pathway and cytokine induction in brown adipocytes
Since the interaction between ASK1 and RIPK2 was originally identified in brown adipocytes, we next investigated the role of ASK1 in the NOD-RIPK2 pathway with an experimental model of brown adipocytes, HIB 1B cells29. As similar with in NOD1-HEK293A cells, ASK1 knockdown in differentiated HIB 1B cells enhanced the K63-polyubiquitination of RIPK2 under NOD1 ligand stimulation (Fig. 3a). Moreover, knockdown of ASK1 in brown adipocytes augmented the C12-iE-DAP-induced degradation of IκBα (Fig. 3b). Note that the maximum effects of ASK1 on the NOD-RIPK2 pathway were observed in different time point between NOD1-HEK293A cells and HIB 1B cells (Fig. 2d,h vs. Fig. 3a,b), implying that the involvement of ASK1 in the NOD-RIPK2 pathway is conserved across cell lines but the NOD-RIPK2 pathway is finetuned differently (e.g., feedback mechanism). We further examined whether the suppressive effects of ASK1 on the NOD-RIPK2 pathway are physiologically relevant to inflammatory cytokine production in brown adipocytes by measuring the mRNA levels of chemokines17,39. ASK1 knockdown exhibited the significant upregulation for the induction of chemokine (C-C motif) ligand 2 (Ccl2, also known as MCP-1), Ccl5 (also known as RANTES), and interleukin 6 (Il6) mRNAs, and the upregulating tendency for tumor necrosis factor (Tnf, typically known as TNFα) and chemokine (C-X-C motif) ligand 2 (Cxcl2) in C12-iE-DAP-treated HIB 1B cells (Fig. 3c). This upregulation of inflammatory cytokines observed under single knockdown of ASK1 was abolished when both ASK1 and RIPK2 were knocked down (Fig. 3d). These results suggest that ASK1 suppresses cytokine production in a NOD-RIPK2-dependent manner.
ASK1 suppresses the NOD-RIPK2 pathway and cytokine induction in brown adipocytes. (a,b) Effects of ASK1 knockdown on the K63-polyubiquitination of RIPK2 (a) or the activation of the NOD-RIPK2 pathway (b) in brown adipocytes. ASK1-knockdown HIB 1B cells were treated with C12-iE-DAP (10 µg/mL) for the indicated times. The right graph in (a) indicates the quantification of western blot, and statistical analysis was performed against Control #2 at each time point. JNK loading control in (b). (c,d) Effects of ASK1 knockdown (c) or ASK1/RIPK2 double knockdown (d) on the relative mRNA levels of inflammatory cytokines under NOD-RIPK2 pathway activation. Differentiated HIB 1B cells were treated with C12-iE-DAP (10 µg/mL, 6 h). (e–g) Effects of ASK1 knockdown on cytokine secretion from NOD-RIPK2 pathway-activated cells. Supernatant (e), or supernatant and cell lysates (g) from C12-iE-DAP (10 µg/mL, 8 h)-treated HIB 1B cells were subjected to cytokine antibody array (e) or immunoblotting (g), respectively. The images of cytokine antibody array were quantified in (f). See Supplementary Fig. S1 for the other cytokines. Data are represented as the mean ± SEM. n = 4 in (a), n = 9 (pooled from 3 independent experiments) in (c), n = 8 (pooled from 4 independent experiments) in (d), n = 6 (pooled from 3 independent experiments) for IL6, CCL2 and n = 8 (pooled from 4 independent experiments) for CCL12, CSF2 in (f). *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant, according to two-tailed Dunnett’s test (a,c) or two-tailed Welch’s t-test with the Bonferroni correction (d,f). Ctrl control.
Subsequently, we investigated the suppressive effect of ASK1 on cytokine production in brown adipocytes at the protein level using a cytokine antibody array targeting 22 mouse cytokines. The systematic approach revealed that stimulation of HIB 1B cells with C12-iE-DAP significantly upregulated IL6, CCL2, CCL5, CCL12, colony stimulating factor 2 (CSF2, also known as GM-CSF), CSF3 (also known as GCSF), IL9, IL12 p40/p70, IL17 and thrombopoietin (also known as THPO) while significantly downregulated soluble TNF receptor-1 (sTNFR1) and vascular endothelial growth factor (VEGF) (Fig. 3e,f, Supplementary Fig. S1), some of which were consistent with the previous report17. Among them, ASK1 knockdown showed significant increase in the C12-iE-DAP-dependent upregulation of IL6 and CSF2 and increasing tendency in that of CCL2 and CCL12 (Fig. 3e,f). Of note, CCL5 was not significantly increased by ASK1 knockdown at the protein level (Supplementary Fig. S1) in contrast to the mRNA result (Fig. 3c), but we cannot rule out the possibility that the effect of ASK1 on CCL5 accumulation can be detected only at later time points due to protein turnover time course of CCL5. Interestingly, the C12-iE-DAP-dependent upregulation of thrombopoietin was significantly decreased by ASK1 knockdown (Supplementary Fig. S1), possibly implying that the effect of ASK1 knockdown on thrombopoietin may be derived via direct ASK1 activity for basal p38/JNK MAPK pathway although we need more detailed analyses. As a representative validation of the cytokine antibody array assay, we confirmed the increased secretion of IL6 by ASK1 knockdown using western blot (Fig. 3g). Altogether, these results suggest that ASK1 suppresses the release of not global but some specific cytokines in brown adipocytes.
ASK1 does not suppress the NOD-RIPK2 pathway and cytokine production in white adipocytes
Mammalian adipocytes are classified into two classes: white adipocytes and brown adipocytes4,5. Our finding that ASK1 suppresses the NOD-RIPK2 pathway in brown adipocytes led us to investigate the involvement of ASK1 in the NOD-RIPK2 pathway in white adipocytes; i.e., we differentiated 3T3-L1 cells13 as an experimental model of white adipocytes and examined the effect of ASK1 knockdown on the NOD-RIPK2 pathway. Stimulation of the differentiated 3T3-L1 cells with C12-iE-DAP activates the NOD-RIPK2 pathway, as indicated by degradation of IκBα (Supplementary Fig. S2a). However, against our expectation, the knockdown of ASK1 did not show the significant enhancement of IκBα degradation (Supplementary Fig. S2a). In addition, while stimulation of 3T3-L1 cells with C12-iE-DAP induced the expression of pro-inflammatory cytokines Ccl2, Ccl5 and Il6 as previously reported13, ASK1 knockdown did not enhance inflammatory cytokine induction under ligand stimulation (Supplementary Fig. S2b). These results indicate that ASK1 regulates the NOD-RIPK2 pathway in a cell type-dependent manner.
Inhibition of the NOD-RIPK2 pathway contributes to maintenance of thermogenic potential in brown adipocytes
Cell type-specific ASK1 suppression implies some physiological meaning of the NOD-RIPK2 pathway in brown adipocytes. Several studies have reported that acute or chronic activation of pattern recognition receptors, namely, Toll-like receptor 2 (TLR2), TLR4 and NOD1, attenuate the expression of brown adipocyte markers in brown adipocytes17,40. Hence, we hypothesized that suppression of the NOD-RIPK2 pathway by ASK1 may contribute to the thermogenic function in brown adipocytes. However, because the PKA-ASK1-p38 axis is involved in the maturation of brown adipocytes19, it may not be easy to distinguish the roles of ASK1 in the NOD-RIPK2- and PKA-p38-dependent regulations of brown adipocytes using a simple knockdown experiment of ASK1 in brown adipocytes. Adipose inflammation is aggravated by local cross-talk between adipocytes and infiltrated macrophages41, and proinflammatory cytokines secreted from macrophages are involved in paracrine regulation of thermogenic function in brown adipocytes42,43. These cytokines can also be secreted from brown adipocytes44. Therefore, we instead established an experimental model to explore a role of ASK1 in the cytokines-mediated regulation of thermogenic potential by measuring β3-adrenergic receptor responsiveness (Supplementary Fig. S3a). Briefly, the inflammatory cytokine-containing culture medium (conditioned medium) was collected from brown adipocytes that were treated with the NOD-RIPK2 pathway activator and/or siRNA against ASK1 in advance (“donor cells”) (cf. Fig. 3e–g). Subsequently, another set of brown adipocytes (“acceptor cells”) was stimulated with a β3-adrenergic receptor agonist following exposure to the conditioned medium, and the induction of brown adipocyte markers in acceptor cells was evaluated. We first examined whether our experimental system could model the inflammatory environments where thermogenic markers are downregulated in brown adipocytes. Compared with control media, conditioned medium from the C12-iE-DAP-treated donor cells significantly suppressed the brown adipocyte markers Ucp1, Prdm16 and Cox4i1 induced by CL316,243, a β3-adrenergic receptor-specific agonist45, in the acceptor HIB 1B cells (Supplementary Fig. S3b), suggesting that our model enables us to evaluate the inflammatory BAT-derived intercellular effects on the thermogenic function of BAT. Thus, we determined the effect of ASK1 knockdown in donor HIB 1B cells on the responsiveness to the β3-adrenergic receptor agonist in acceptor cells. ASK1 knockdown in donor HIB 1B cells aggravated the inhibitory effect of C12-iE-DAP-treated conditioned medium on brown adipocyte markers upon CL316,243 administration in acceptor HIB 1B cells (Supplementary Fig. S3c). Altogether, our results support the hypothesis that the inhibitory effect of ASK1 on the NOD-RIPK2 pathway is involved in maintaining the thermogenic potential of brown adipocytes in an inflammatory environment.
Discussion
In this study, we established a novel chemical pull-down MS method and identified RIPK2 as an ASK1 interactor in brown adipocytes. The affinity purification-MS (AP-MS) method has been one of the representative footholds to characterize the regulations and functions of a protein of interest, and we have indeed conducted the AP-MS analyses using samples of tagged-ASK1-overexpressing HEK293A cells27,46. However, none of the previous trials identified RIPK2 as an ASK1 interactor. Although purification of overexpressed protein is most commonly used in AP-MS, the method often faces several issues. For instance, tagging at the terminus of a protein may affect the conformation or subcellular localization of the protein and impede the access of its binding partners47, which reduces the protein interactions in cells and also in solution through pull-down step. Overexpressed proteins can also interact with artificial partners in cells, which makes it difficult to distinguish genuine endogenous interactors. In addition, a strong affinity between avidin and biotin (KD ~ 10−15 [M]), one of the most commonly used combinations for chemical pull-down systems, makes it difficult to elute the protein complex without the alteration of pH or temperature or the addition of denaturants48, which is not optimal for elution condition. Besides, purification of endogenous protein complexes depends largely on the availability of antibodies for pull-down assays; thus, there have been only a few reports on identifying components of endogenous signalosomes. We propose that our novel ASKA pull-down MS method overcomes major drawbacks in the typical AP-MS methods and hence is a powerful AP-MS option that is applicable to a broad range of endogenous kinases when identifying genuine components of its signalosome. To utilize the high specificity of 1NA-PP1 to the as-kinase, ASKA technology introduces mutations in the ATP-binding pockets22,49. The structure and sequence of the ATP-binding pocket are so highly conserved that this kinase modification methodology has been applied to numerous kinases. The recent advancement in genome editing technology (e.g., CRISPR-Cas9 system50) provides an easier environment to introduce gene modification. Previous reports proved that ASKA modification did not considerably affect the ATP-binding ability or kinase activity of the as-kinase23,51, and we also demonstrated that purification of as-kinase with 1NA-PP1 did not significantly alter the total size of the kinase signalosome (Fig. 1g). We believe the ASKA pull-down MS method will facilitate protein–protein interaction analyses of any desired kinase, and is highly promising as a tool to drive kinase research.
BAT is a unique organ for energy expenditure through thermogenesis. Inflammation in brown adipocytes is of great research interest but remains relatively unknown compared to white adipose inflammation. In this study, we showed that ASK1 suppresses the inflammatory NOD-RIPK2 pathway and cytokine secretion in brown adipocytes. While ASK1 has been reported as a critical regulator of inflammation23,34,52, most of these reports demonstrated that ASK1 promotes the inflammatory response. Our finding is unique in that ASK1 suppresses the inflammatory response. A remaining question lies in the detailed regulatory mechanism of the NOD-RIPK2 pathway by ASK1. The simplest explanation would be that ASK1 physically hinders the modification of RIPK2 or recruitment of its downstream effector molecules. We showed that ASK1 specifically bound to the kinase domain of RIPK2 (Fig. 2f) and inhibited the recruitment of an E3 ubiquitin ligase XIAP and the K63-polyubiquitination of RIPK2 (Fig. 2g,h). The kinase domain of RIPK2 (18–290 amino acids; Fig. 2e) harbors several important residues involved in downstream activation. For instance, S176 in the activation loop is autophosphorylated under NOD ligand stimulation53. K63-type polyubiquitination on K209 is important for the recruitment of the TAB/TAK1 complex and the subsequent activation of downstream NF-κB signaling36. The binding interface between XIAP and RIPK2 was mapped in the kinase domain of RIPK254. Hence, binding of ASK1 to the kinase domain might function as a direct physical obstruction or induce structural change of RIPK2 to block these modifications.
As a potential biological significance of the ASK1-dependent RIPK2 regulation in BAT, our in vitro model with HIB 1B cells suggested that the suppression of inflammation in brown adipocytes by ASK1 contributes to the intercellular maintenance of thermogenic capacity in brown adipocytes (Supplementary Fig. S3). The result is in accordance with our previous report that ASK1 is involved in brown adipocyte maturation via activation of the PKA-ASK1-p38 axis19; that is, our studies underpin the multifaceted regulatory mechanism of brown adipocyte maintenance by ASK1 (Fig. 4). It is reasonable that a single signaling molecule controls the same physiological function under different situations—in this case, thermogenic gene expression under normal differentiation processes and under inflammation. However, we should still keep the limitations of our in vitro model in mind. For instance, although adipose inflammation is aggravated by local cross-talk between adipocytes and immune cells in the whole body, our in vitro model does not take the effects of immune cells into account. Moreover, we tested our hypothesis using HIB 1B cells. Although utilized as a brown adipocyte cell line, HIB 1B cells express limited amount of brown adipocyte markers such as Ucp1 and β3-adrenergic receptor compared to primary brown adipocytes derived from rodents29. Further studies with primary culture cells and in vivo systems will be needed to validate our hypothesis and test the biological effect size in vivo.
Hypothetical model. Through interacting with RIPK2, ASK1 negatively regulates the NOD-RIPK2 pathway and inflammatory cytokine production in brown adipocytes. Along with the maturation-enhancing effect of ASK1 via the PKA-ASK1-p38 axis under β3-adrenergic receptor stimulation19, this regulation would contribute to maintaining brown adipocyte function under inflammation.
As another clue for the biological significance of our findings, we found that ASK1 does not suppress the NOD-RIPK2 pathway in white adipocytes (Supplementary Fig. S2). Activation of the NOD-RIPK2 pathway in white adipocytes induces insulin resistance, that is, it seems to be maladaptive for energy homeostasis13,15. However, inflammation in white adipocytes can also have a beneficial effect on promoting WAT expansion and remodeling, which limits the permeability of bacterial fragments as an intestinal barrier55. Hence, adaptive reprogramming in WAT against increased energy uptake55 may be blocked by inhibition of the NOD-RIPK2 signaling in white adipocytes. On the other hand, inflammation in BAT suppresses UCP1 expression in brown adipocytes and thus limits the energy expenditure under inflammation17. BAT-specific regulation of the NOD-RIPK2 pathway by ASK1 may contribute to effectively maintaining the thermogenic function of brown adipocytes without impairing the inflammation-driven reprogramming of WAT. The molecular mechanism of how ASK1 achieves brown adipocyte-specific regulation of the NOD-RIPK2 pathway still needs future investigation.
From a macro perspective, adipose inflammation is a crucial hub for obesity and metabolic dysregulation. Chronic low-grade inflammation of adipose tissue, characterized by increased secretion of inflammatory cytokines and infiltration of macrophages and other types of immune cells, is observed under obesity and regarded as a trigger of metabolic disorders, including type 2 diabetes and cardiovascular diseases3. The physiological ligand of NODs, peptidoglycan, is considered to be derived from gut microbiota and translocated from the luminal side of the mucosa into the host circulation11. High-fat diet feeding affects gut microbiota and enhances intestinal permeability12,56. Besides, Nod1 and Nod2 double knockout mice are protected from high fat diet-induced insulin intolerance15, and several reports pointed out that the NOD-RIPK2 pathway is activated in adipose tissue from patients with metabolic syndrome or diabetes57,58. These lines of evidence imply that obesity may facilitate inflammation and metabolic dysregulation in a NOD-RIPK2-dependent manner. In this study, we showed that ASK1 downregulated the NOD-RIPK2 pathway (Fig. 3, Supplementary Fig. S3), which suggests the potential function of ASK1 as an adipose inflammation suppressor by regulating the NOD-RIPK2 pathway. At the same time, ASK1 expression in adipose tissue is upregulated under obesity both in human59,60 and in mice61, and kinase activity of ASK1 is upregulated in mice under high fat diet-feeding62, which rather propose that ASK1 expression and/or activation promote adipose inflammation. Alternatively, our results may point out the existence of latent beneficial aspect of ASK1 upregulation through tuning the NOD-RIPK2 pathway under obesity. As a potential relevant finding to this assumption, we previously demonstrated that global Ask1 knockout mice showed impaired glucose clearance compared to wild-type mice under high-fat diet treatment, and the phenotype was more robust in severely obese animals20. Unexpectedly, however, adipocyte-specific Ask1 knockout mice exhibited comparable glucose clearance with wild-type20, which denies the potential influence of ASK1 on blood glucose level under obesity through downregulating the pro-inflammatory NOD-RIPK2 pathway in adipocytes. Nevertheless, considering the confounding example of body weight in the phenotype of Ask1 knockout mice and complexity of obesity-induced inflammatory response in vivo, we should keep in mind that negative regulation of the NOD-RIPK2 pathway by ASK1 in brown adipocytes may contribute to unidentified whole-body phenotype in some obesity-associated situation.
In conclusion, our work demonstrated an example application of the ASKA pull-down MS method and revealed a novel regulatory mechanism in adipose inflammation that ASK1 suppresses inflammatory NOD-RIPK2 signaling in brown adipocytes. As a crucial hub of adipose inflammation and thermogenic gene induction, the NOD-RIPK2 pathway in brown adipocytes would be an attractive therapeutic target against obesity-associated diseases. Further investigation of the regulatory mechanism of the NOD-RIPK2 pathway by ASK1 may provide new insight into the clinical approach to obesity.
Methods
Antibodies and reagents
Antibodies used in this study are listed in Supplementary Table S163.
C12-iE-DAP (#tlrl-c12dap) was purchased from InvivoGen, dissolved at 10 mg/mL in dimethyl sulfoxide (DMSO; Sigma-Aldrich, #D5879) and dissolved at a final concentration of 1000× in LAL water (InvivoGen, #h2olal-1.5). CL316,243 (#sc-203895) was purchased from Santa Cruz Biotechnology, and H2O2 (#081-04215) was purchased from Wako Pure Chemical Industries and dissolved at a final concentration of 1000× in ultrapure water. Cycloheximide was purchased from Sigma-Aldrich and dissolved at a final concentration of 100 mg/mL in DMSO.
Cell lines and cell culture
HEK293A cells (Invitrogen) were cultured in DMEM-high glucose (Sigma-Aldrich, #D5796) supplemented with 10% fetal bovine serum (FBS, biowest). HIB 1B cells, a kind gift from Dr. Bruce M. Spiegelman (Harvard Medical School and Dana-Farber Cancer Institute), were cultured in DMEM-high glucose supplemented with 10% FBS. Of note, to eliminate mycoplasma, the original HIB 1B cells were treated with MynoxGold (Minerva Biolabs, #10-0201) and subcloned by limiting dilution. 3T3-L1 cells, a kind gift from Dr. Shin-Ichiro Takahashi (The University of Tokyo), were cultured in DMEM-high glucose supplemented with 10% calf serum (Sigma-Aldrich, #C8056) and 100 units/mL penicillin G (Meiji Seika, #6111400D2039). Tetracycline-inducible mNOD1-6Myc-stably expressing HEK293A (NOD1-HEK293A) cells were established as previously described64 and cultured in DMEM-high glucose supplemented with 10% FBS, 2.5 µg/mL blasticidin (Invitrogen, #A1113903), and 50 µg/mL Zeocin (Invitrogen, #R25001). To induce mNOD1-6Myc, cells were pretreated with 1 μg/mL tetracycline (Sigma-Aldrich, #T7660) 1 day before assays.
For brown adipocyte differentiation, HIB 1B cells were plated at 1.0 × 105 cells/mL on day 4. Cells were exposed to differentiation induction medium (DMEM-high glucose containing 1 nM triiodo-l-thyronine (T3; Sigma-Aldrich, #T2877), 20 nM insulin (Sigma-Aldrich, #I1882), 5 µM dexamethasone (Sigma-Aldrich, #D4902), 0.125 mM indomethacin, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX; Sigma-Aldrich, #I5879), 1 µM rosiglitazone (Sigma-Aldrich, #R2408), 20 mM HEPES–NaOH (pH 7.4), 20% FBS) on day 0. The medium was changed to differentiation enhancement medium (DMEM-high glucose containing 1 nM T3, 20 nM insulin, 20 mM HEPES–NaOH (pH 7.4), 20% FBS) on days 2, 4 and 6. The medium was replaced with fresh DMEM-high glucose supplemented with 20% FBS 1 h prior to starting experiments.
For white adipocyte differentiation, 3T3-L1 cells were plated at 1.0 × 105 cells/mL on day 4. Cells were exposed to differentiation induction medium (DMEM-high glucose containing 1.7 mM insulin, 250 nM dexamethasone, 0.5 mM IBMX, 10% FBS) on day 0. The medium was changed to differentiation enhancement medium (DMEM-high glucose containing 1.7 mM insulin, 10% FBS, DE) on days 2, 4, and 6, resuspended to 5.0 × 105 cells/mL on day 8 with DE, and replaced with fresh DE on day 10. The medium was replaced with fresh DMEM-high glucose supplemented with 10% FBS 1 h prior to starting the experiments.
Primary brown fat stromal vascular fraction (SVF) was isolated from newborn Ask1ASKA knock-in mice and differentiated into brown adipocytes as previously reported19. All experiments were performed in accordance with protocols approved by the Animal Research Committee of the Graduate School of Pharmaceutical Sciences, The University of Tokyo (Tokyo, Japan).
In silico analysis of the ASK1 ATP-binding pocket
The previously reported crystal structure of the ASK1 kinase domain (PDB: 2CLQ)24 was analyzed using AutoDock 4.0 (Scripps Research) software to calculate the depth of the ATP-binding pocket.
Synthesis of 1NA-PP1 derivatives
1NA-PP1-L1 and 1NA-PP1-L2 were synthesized as described in Supporting Information (Supplementary Note)65,66.
Mass spectrometry analysis
The as-ASK1 complex was purified from primary brown adipocytes (day 4). After concentration by TCA precipitation, the purified as-ASK1 samples were dissolved in guanidine hydrochloride and digested with lysyl endopeptidase (Lys-C; Wako Chemicals USA) and then further digested with trypsin (Sigma-Aldrich). All samples were analyzed by a direct nanoflow liquid chromatography (LC) system coupled to a time-of-flight mass spectrometer (Triple TOF 5600+; AB Sciex) as previously described67. We regarded the identified protein as an interactor candidate of ASK1: (1) if the protein was uniquely mapped from the obtained peptides and (2) if the number of peptides obtained from 1NA-PP1-eluted samples was larger than that from DMSO-eluted negative control samples.
In a meta-analysis of different pull-down MS results, the identified mouse proteins in the ASKA pull-down MS method were converted to human orthologs using the NCBI Entrez database in a highly conservative manner; two proteins, Pcdhgb8 and Vmn1r87, were unable to be assigned to human orthologs. For the Flag-tag pull-down MS result27, we set criteria for interactor candidates of ASK1: (1) if the protein was uniquely mapped from the obtained peptides and (2) if the number of peptides obtained from the wild-type ASK1-transfected samples was larger than that from mock negative control samples. Regarding the other Flag-tag pull-down MS result28, we regarded the listed protein as an interactor candidate of ASK1 if the significance analysis of interactome (SAINT) score was more than 0.6.
Expression plasmids
Expression plasmids for this study were constructed by standard molecular biology techniques, and all constructs were verified by sequencing. A mouse ASK1 cDNA (CDS of NM_008580.4 harboring four silent mutants in its nucleotide sequence and K203R mutant and lacking stop codon) was previously cloned and subcloned into pcDNA3 (Invitrogen) with a C-terminal HAsl tag46. cDNAs of mouse RIPK2 (CDS of NM_138962.4), mouse NOD1 (CDS of NM_172729.3), and mouse XIAP (CDS of NM_001301639.1) were subcloned into pcDNA3/GW or pcDNA4 (Invitrogen) with an N- or C-terminal Flag, a C-terminal 6Myc, or an N-terminal YFP. RIPK2 mutants KD (CDS of NM_138962.4 with c.874_1,617del), IM (CDS of NM_138962.4 with c.1_873del, c.1291_1617del), and CARD (CDS of NM_138962.4 with c.1_1302del) were constructed from full-length RIPK2 and subcloned into pcDNA3/GW with an N-terminal Flag and C-terminal YFP.
Transfections
Plasmid transfections were performed with polyethylenimine “MAX” (Polysciences, #24,765) when HEK293A cells were grown to 80–95% confluency, according to a previously described protocol68 with minor optimization. In certain experiments, after 4–6 h of transfection, the cells were cultured in fresh medium for another 40 h to reduce cytotoxicity.
siRNA transfections were carried out by reverse transfection using Lipofectamine RNAiMAX (Invitrogen, #133778-150) according to the manufacturer’s instructions. For siRNA transfection of NOD1-HEK293A cells, cells were reverse transfected with 10 nM siRNA on the day of seeding. For siRNA transfection of HIB 1B cells, cells were resuspended in DE buffer and transfected with 16.7 nM siRNAs on day 4 or resuspended in DI buffer on day 0 and in DE buffer on day 4 with reverse transfection of 16.7 nM siRNAs. For siRNA transfection of 3T3-L1 cells, cells were resuspended in DE buffer and transfected with 4.17 nM siRNAs on day 8. siRNA sequences are listed in Supplementary Table S2.
Cell lysis and immunoblotting
Cells were lysed in lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10 mM EDTA, 1% sodium deoxycholate, and 1% Triton X-100) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 μg/mL leupeptin, and phosphatase inhibitor cocktail (20 mM NaF, 30 mM β-glycerophosphatase, 2.5 mM Na3VO4, 3 mM Na2MoO4, 12.5 μM cantharidin and 5 mM imidazole). For the samples in Fig. 1I, cells were lysed in lysis buffer containing 10 mM N-ethylmaleimide (Tokyo Chemical Industry, #E0136) dissolved in ethanol (Wako Pure Chemicals Industries, #057000451). Cell extracts were clarified by centrifugation, and supernatants were sampled by adding SDS sample buffer. In certain experiments, 5% volume/volume β-mercaptoethanol (Nacalai Tesque, #21438082) was added to SDS sample buffer.
For the supernatant in Fig. 3g, culture medium was substituted into medium supplemented with 0.2% FBS prior to experiments. Culture medium was collected and centrifuged for 5 min to separate the supernatant from a pellet. The supernatants were mixed with fourfold volumes of ice-cold acetone (Wako Pure Chemicals Industries, #012-00343), followed by incubation at − 20 °C over 2 h and subsequent sedimentation through centrifugation. The samples were dissolved in SDS sample buffer.
After boiling at 98 °C for 3 min, the samples were resolved by SDS-PAGE and electroblotted onto a BioTrace PVDF membrane (Pall) or Immobilon-P membrane (Millipore, #IPVH00010). The membranes were blocked with 2% skim milk (Megmilk Snow Brand) in TBS-T (20 mM Tris–HCl pH 8.0, 137 mM NaCl and 0.1% Tween 20) and probed with the appropriate primary antibodies diluted by 1st antibody-dilution buffer (TBS-T supplemented with 5% BSA (Iwai Chemicals, #A001) and 0.1% NaN3 (Nacalai Tesque, #312-33)). After replacing and probing the appropriate secondary antibodies diluted with skim milk in TBS-T, antibody-antigen complexes were detected on X-ray films (FUJIFILM, #47410-07523, #47410-26615 or #47410-07595) using an ECL system (GE Healthcare). Quantification was performed against the raw digital images with densitometry using Fiji/ImageJ software69.
Cytokine antibody array
Cytokines secreted into culture medium were quantified using Mouse Cytokine Antibody Array (Membrane, 22 targets) (abcam, #ab133993) following the manufacturer’s protocol. Briefly, culture medium was substituted into fresh medium supplemented with 0.2% FBS prior to experiments, and culture medium was collected 8 h after the C12-iE-DAP stimulation (10 µg/mL). 100-fold diluted culture medium was incubated with an anti-cytokine antibody-arrayed membrane overnight at 4 °C. The target cytokines-trapped membrane was further incubated with the biotin-conjugated anti-cytokine antibody overnight at 4 °C, followed by the reaction with horseradish peroxidase (HRP)-streptavidin. The HRP-labelled cytokines was detected with chemiluminescence using a chemiluminescence imaging system FUSION SOLO.7S.EDGE (Vilber). Because the detection range in this kit is dependent on each cytokine, the detected membrane was adequately washed with TBS-T and iterated the above detection procedure for the undiluted culture medium.
The obtained images were adjusted by rolling ball background subtraction and quantified with using ImageJ/Fiji software. The quantified values were scaled with the values of negative controls and positive controls in each membrane.
Statistical analysis
The data are summarized as the mean ± SEM. No statistical method was utilized to predetermine the sample size. Statistical tests, the number of samples and the sample sizes are indicated in each figure legend. Statistical tests were performed using R with RStudio software, and P < 0.05 was considered statistically significant. The investigators were not blinded to allocation during experiments and outcome assessments. The experiments were not randomized. However, the experiments were performed across different passages of cells, and the cells in the control and treated groups were seeded from the same population of cells.
Other methods in this work, including surface plasmon resonance assay, gel filtration column chromatography, preparation of 1NA-PP1-Lx-immobilized beads, purification of the as-ASK1 signalosome, quantitative RT-PCR analysis, coimmunoprecipitation assay and TUBE pull-down assay, are described in Supplementary Methods. All methods are reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org), but further information and requests for resources and reagents should be directed to K.W. and H.I.
Data availability
The authors declare that all data supporting the findings of this study are available within the main article or the supplementary materials. Source data are provided with this paper as Supplementary Data.
Change history
29 November 2021
The original online version of this Article was revised: In the original version of this Article the ORCID ID for Tomoya Hirano was incorrectly assigned to Tomohisa Hatta. The ORCID ID for Tomoya Hirano is 0000-0003-1196-7199.
References
Schäffler, A. & Schölmerich, J. Innate immunity and adipose tissue biology. Trends Immunol. 31, 228–235 (2010).
Villarroya, F., Cereijo, R., Villarroya, J. & Giralt, M. Brown adipose tissue as a secretory organ. Nat. Rev. Endocrinol. 13, 26–35 (2017).
Reilly, S. M. & Saltiel, A. R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13, 633–643 (2017).
Chouchani, E. T., Kazak, L. & Spiegelman, B. M. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 29, 27–37 (2019).
Chouchani, E. T. & Kajimura, S. Metabolic adaptation and maladaptation in adipose tissue. Nat. Metab. 1, 189–200 (2019).
Villarroya, F., Cereijo, R., Gavaldà-Navarro, A., Villarroya, J. & Giralt, M. Inflammation of brown/beige adipose tissues in obesity and metabolic disease. J. Intern. Med. 284, 492–504 (2018).
Alcalá, M. et al. Increased inflammation, oxidative stress and mitochondrial respiration in brown adipose tissue from obese mice. Sci. Rep. 7, 1–12 (2017).
Sakamoto, T. et al. Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol. Metab. 310, E676–E687 (2016).
Moreira, L. O. & Zamboni, D. S. NOD1 and NOD2 signaling in infection and inflammation. Front. Immunol. 3, 328 (2012).
Heim, V. J., Stafford, C. A. & Nachbur, U. NOD signaling and cell death. Front. Cell Dev. Biol. 7, 208 (2019).
Clarke, T. B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Cani, P. D. et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481 (2008).
Zhao, L., Hu, P., Zhou, Y., Purohit, J. & Hwang, D. NOD1 activation induces proinflammatory gene expression and insulin resistance in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 301, 587–598 (2011).
Chi, W. et al. Bacterial peptidoglycan stimulates adipocyte lipolysis via NOD1. PLoS ONE 9, 1–9 (2014).
Schertzer, J. D. et al. NOD1 activators link innate immunity to insulin resistance. Diabetes 60, 2206–2215 (2011).
Purohit, J. S. et al. The effects of NOD activation on adipocyte differentiation. Obesity 21, 737–747 (2012).
Bae, J. et al. Activation of pattern recognition receptors in brown adipocytes induces inflammation and suppresses uncoupling protein 1 expression and mitochondrial respiration. Am. J. Physiol. Physiol. 306, C918–C930 (2014).
Ichijo, H. et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science 275, 90–94 (1997).
Hattori, K. et al. ASK1 signalling regulates brown and beige adipocyte function. Nat. Commun. 7, 1–10 (2016).
Hattori, K. et al. β-adrenergic receptor signaling evokes the PKA-ASK axis in mature brown adipocytes. PLoS ONE 15, e0232645 (2020).
Noguchi, T. et al. Recruitment of tumor necrosis factor receptor-associated factor family proteins to apoptosis signal-regulating kinase 1 signalosome is essential for oxidative stress-induced cell death. J. Biol. Chem. 280, 37033–37040 (2005).
Bishop, A. C. et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 (2000).
Mizukami, J. et al. ASK1 promotes the contact hypersensitivity response through IL-17 production. Sci. Rep. 4, 1–8 (2014).
Bunkoczi, G. et al. Structural and functional characterization of the human protein kinase ASK1. Structure 15, 1215–1226 (2007).
Takeda, K. et al. Apoptosis signal-regulating kinase (ASK) 2 functions as a mitogen-activated protein kinase kinase kinase in a heteromeric complex with ASK1. J. Biol. Chem. 282, 7522–7531 (2007).
Wang, H., Fan, L., Wang, H., Ma, X. & Du, Z. Amyloid β regulates the expression and function of AIP1. J. Mol. Neurosci. 55, 227–232 (2014).
Takeda, K. et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc. Natl. Acad. Sci. U.S.A. 106, 12301–12305 (2009).
Federspiel, J. D. et al. Assembly dynamics and stoichiometry of the apoptosis signal-regulating kinase (ASK) signalosome in response to electrophile stress. Mol. Cell. Proteomics 15, 1947–1961 (2016).
Klaus, S. et al. Characterization of the novel brown adipocyte cell line HIB 1B. Adrenergic pathways involved in regulation of uncoupling protein gene expression. J. Cell Sci. 107, 313–319 (1994).
Golkowski, M. et al. Kinobead and single-shot LC-MS profiling identifies selective PKD inhibitors. J. Proteome Res. 16, 1216–1227 (2017).
Humphries, F., Yang, S., Wang, B. & Moynagh, P. N. RIP kinases: Key decision makers in cell death and innate immunity. Cell Death Differ. 22, 225–236 (2015).
Lee, J. et al. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J. Biol. Chem. 284, 23818–23829 (2009).
Saitoh, M. et al. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606 (1998).
Matsuzawa, A. et al. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat. Immunol. 6, 587–592 (2005).
Inohara, N., Del Peso, L., Koseki, T., Chen, S. & Núñez, G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem. 273, 12296–12300 (1998).
Hasegawa, M. et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 27, 373–383 (2008).
Krieg, A. et al. XIAP mediates NOD signaling via interaction with RIP2. Proc. Natl. Acad. Sci. U.S.A. 106, 14524–14529 (2009).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
Canning, P. et al. Inflammatory signaling by NOD-RIPK2 is inhibited by clinically relevant type II kinase inhibitors. Chem. Biol. 22, 1174–1184 (2015).
Bae, J., Chen, J. & Zhao, L. Chronic activation of pattern recognition receptors suppresses brown adipogenesis of multipotent mesodermal stem cells and brown pre-adipocytes. Biochem. Cell Biol. 93, 251–261 (2015).
Villarroya, F., Cereijo, R., Villarroya, J., Gavaldà-Navarro, A. & Giralt, M. Toward an understanding of how immune cells control brown and beige adipobiology. Cell Metab. 27, 954–961 (2018).
Goto, T. et al. Proinflammatory cytokine interleukin-1β suppresses cold-induced thermogenesis in adipocytes. Cytokine 77, 107–114 (2016).
Sakamoto, T. et al. Inflammation induced by RAW macrophages suppresses UCP1 mRNA induction via ERK activation in 10T1/2 adipocytes. Am. J. Physiol. Cell Physiol. 304, C729–C738 (2013).
Omran, F. & Christian, M. Inflammatory signaling and brown fat activity. Front. Endocrinol. 11, 156 (2020).
Bloom, J. D. et al. Disodium (R, R)-5-[2-[[2-(3-chlorophenyl)-2-hydroxyethyl]amino]propyl]-1,3-benzodioxole-2,2-dicarboxylate (cl 316,243). A potent β-adrenergic agonist virtually specific for β3 receptors. A promising antidiabetic and antiobesity agent. J. Med. Chem. 35, 3081–3084 (1992).
Cheng, R. et al. β-TrCP-dependent degradation of ASK1 suppresses the induction of the apoptotic response by oxidative stress. Biochim. Biophys. Acta Gen. Subj. 1862, 2271–2280 (2018).
Morris, J. H. et al. Affinity purification–mass spectrometry and network analysis to understand protein-protein interactions. Nat. Protoc. 9, 2539–2554 (2014).
Rösli, C., Rybak, J.-N., Neri, D. & Elia, G. Quantitative recovery of biotinylated proteins from streptavidin-based affinity chromatography resins. In Avidin-Biotin Interactions (ed. McMahon, R. J.) 89–100 (Humana Press, 2008).
Zhang, C. et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods 2, 435–441 (2005).
Hsu, P. D., Lander, E. S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Liu, Y., Shah, K., Yang, F., Witucki, L. & Shokat, K. M. Engineering Src family protein kinases with unnatural nucleotide specificity. Chem. Biol. 5, 91–101 (1998).
Yoshikawa, M. et al. ASK1 promotes uterine inflammation leading to pathological preterm birth. Sci. Rep. 10, 1–11 (2020).
Dorsch, M. et al. Identification of a regulatory autophosphorylation site in the serine-threonine kinase RIP2. Cell. Signal. 18, 2223–2229 (2006).
Hrdinka, M. et al. Small molecule inhibitors reveal an indispensable scaffolding role of RIPK 2 in NOD 2 signaling. EMBO J. 37, 1–16 (2018).
Wernstedt Asterholm, I. et al. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 20, 103–118 (2014).
Cani, P. D. et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 (2007).
Zhou, Y. J. et al. Increased NOD1, but not NOD2, activity in subcutaneous adipose tissue from patients with metabolic syndrome. Obesity 23, 1394–1400 (2015).
Lappas, M. NOD1 expression is increased in the adipose tissue of women with gestational diabetes. J. Endocrinol. 222, 99–112 (2014).
Blüher, M. et al. Activated Ask1-MKK4-p38MAPK/JNK stress signaling pathway in human omental fat tissue may link macrophage infiltration to whole-body insulin sensitivity. J. Clin. Endocrinol. Metab. 94, 2507–2515 (2009).
Haim, Y. et al. ASK1 (MAP3K5) is transcriptionally upregulated by E2F1 in adipose tissue in obesity, molecularly defining a human dys-metabolic obese phenotype. Mol. Metab. 6, 725–736 (2017).
Lucchini, F. C. et al. ASK1 inhibits browning of white adipose tissue in obesity. Nat. Commun. 11, 1642 (2020).
Xiang, M. et al. Targeting hepatic TRAF1-ASK1 signaling to improve inflammation, insulin resistance, and hepatic steatosis. J. Hepatol. 64, 1365–1377 (2016).
Tobiume, K. et al. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2, 222–228 (2001).
Watanabe, K., Umeda, T., Niwa, K., Naguro, I. & Ichijo, H. A PP6-ASK3 module coordinates the bidirectional cell volume regulation under osmotic stress. Cell Rep. 22, 2809–2817 (2018).
Kim, Y. S., Kim, K. M., Song, R., Jun, M. J. & Sohn, Y. S. Synthesis, characterization and antitumor activity of quinolone-platinum(II) conjugates. J. Inorg. Biochem. 87, 157–163 (2001).
Murphy, R. C. et al. Discovery of potent and selective inhibitors of CDPK1 from C. parvum and T. gondii. ACS Med. Chem. Lett. 1, 331–335 (2010).
Chino, H., Hatta, T., Natsume, T. & Mizushima, N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell 74, 909-921.e6 (2019).
Longo, P. A., Kavran, J. M., Kim, M. S. & Leahy, D. J. Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 (2013).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank Dr. Bruce M. Spiegelman (Harvard Medical School and Dana-Farber Cancer Institute) for kindly providing HIB 1B cells; Dr. Shin-Ichiro Takahashi (The University of Tokyo) for kindly providing 3T3-L1 cells; and all members of the Laboratory of Cell Signaling for fruitful discussion.
Funding
This work was supported by the Japan Agency for Medical Research and Development (AMED) under the Project for Elucidating and Controlling Mechanisms of Aging and Longevity (Grant Number JP21gm5010001 to H.I.), by the Japan Society for the Promotion of Science (JSPS) under the Grants-in-Aid for Scientific Research (KAKENHI; Grant Numbers JP19K16067 to K.W., JP16K18872 to K.H., JP18H02569 to I.N. and JP25650061 and JP21H04760 to H.I.) and by the Japan Science and Technology Agency (JST) Moonshot R&D—MILLENNIA Program (Grant Number JPMJMS2022-18 to H.I.).
Author information
Authors and Affiliations
Contributions
S.T., K.W., T.M., K.H. and H.I. Conceptualization; S.T., T.M., M.O., K.M., M.S. and K.H. Data curation; S.T. Formal analysis; K.W., K.H., I.N. and H.I. Funding acquisition; S.T., T.M., M.O., K.M., M.S., T.Hatta, T.N. and K.H. Investigation; S.T., T.M., M.O. and K.H. Methodology; K.W., T.M., K.H. and H.I. Project administration; S.T., M.S., T.Hirano and H.K. Resources; K.W., T.M., K.H. and H.I. Supervision; S.T. Validation; S.T., K.W. and T.Hirano Visualization; S.T., K.W., K.H., I.N. and H.I. Writing—original draft; S.T., K.W., K.H., I.N. and H.I. Writing—review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Takayanagi, S., Watanabe, K., Maruyama, T. et al. ASKA technology-based pull-down method reveals a suppressive effect of ASK1 on the inflammatory NOD-RIPK2 pathway in brown adipocytes. Sci Rep 11, 22009 (2021). https://doi.org/10.1038/s41598-021-01123-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01123-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.