Abstract
The majority of genes encoding photosynthesis-associated proteins in the nucleus are induced by light during photomorphogenesis, allowing plants to establish photoautotrophic growth. Therefore, optimizing the protein import apparatus of plastids, designated as the translocon at the outer and inner envelope membranes of chloroplast (TOC–TIC) complex, upon light exposure is a prerequisite to the import of abundant nuclear-encoded photosynthesis-associated proteins. However, the mechanism that coordinates the optimization of the TOC–TIC complex with the expression of nuclear-encoded photosynthesis-associated genes remains to be characterized in detail. To address this question, we investigated the mechanism by which plastid protein import is regulated by light during photomorphogenesis in Arabidopsis. We found that the albino plastid protein import2 (ppi2) mutant lacking Toc159 protein import receptors have active photoreceptors, even though the mutant fails to induce the expression of photosynthesis-associated nuclear genes upon light illumination. In contrast, many TOC and TIC genes are rapidly induced by blue light in both WT and the ppi2 mutant. We uncovered that this regulation is mediated primarily by cryptochrome 1 (CRY1). Furthermore, deficiency of CRY1 resulted in the decrease of some TOC proteins in vivo. Our results suggest that CRY1 plays key roles in optimizing the content of the TOC–TIC apparatus to accommodate the import of abundant photosynthesis-associated proteins during photomorphogenesis.
Similar content being viewed by others
Introduction
Chloroplasts are organelles found in photosynthetic tissues of plants, and are thought to have originated from a cyanobacterium engulfed by a eukaryotic cell1. Most of the genes that are encoded by the cyanobacterial ancestor have been transferred to the host nuclear genome during evolution. Therefore, the expression of nuclear genes encoding chloroplast proteins, and the import of those proteins into chloroplasts are indispensable for chloroplast development. Protein import from the cytosol to chloroplasts is primarily mediated by the translocon at the outer and inner envelope membranes of chloroplast (TOC–TIC) complexes2,3,4,5. In Arabidopsis thaliana (Arabidopsis), the core TOC complex includes families of Toc159, Toc33 and Toc75 proteins2. Among them, Toc75 constitutes the protein-conducting channel, and Toc159 and Toc34 families possess a GTPase domain and serve as chloroplast precursor protein receptors2,3,6. Although the role of each TIC component is still controversial, there is a consensus that Tic20 serves as the pore of the TIC complex7,8,9,10. As such, TOC–TIC complexes play key roles in delivering nuclear-encoded proteins into chloroplasts.
Genes encoding photosynthesis-associated proteins, designated as photosynthesis-associated nuclear genes (PhANGs), are strongly induced by light11,12. This necessitates a close relationship between the status of the chloroplast protein import system and the amount of photosynthesis-associated proteins to be imported upon light illumination. For example, light induction of PhANGs does not occur in the plastid protein import2 (ppi2) mutant of Arabidopsis lacking Toc159, a major protein import receptor of photosynthesis-associated proteins13. The interpretation of this phenomenon has been that defective plastids send retrograde signals to suppress the expression of PhANGs, thereby preventing the accumulation of unimported precursor proteins in the cytosol13,14,15. On the other hand, the chromophore of phytochromes, phytochromobilin, is synthesized in plastids16,17. It has been well known that phytochromes up-regulate a number of PhANGs upon light exposure18,19. Hence, one can also argue that ppi2 does not possess sufficient functional phytochromes to induce the expression of PhANGs. However, little is known about the mechanism by which plastid protein import is coordinated with the expression of PhANGs.
One possible mechanism that may link plastid protein import and the expression of PhANGs is light-regulated expression of TOC and TIC genes. Several studies have shown that the expression of TOC and TIC genes is subject to developmental regulation. Among the TOC159 family, TOC159 is highly abundant in photosynthetic green tissues, and less abundant in etiolated tissues and roots20,21,22,23. This is consistent with the proposal that Toc159 is the major protein import receptor for photosynthesis-associated proteins. Likewise, the expression of TOC33 and TOC34 is much higher in light-grown plants than in dark-grown plants24. TIC40 and TIC110 are more abundant in leaves than in roots22, and this is consistent with the observation that leaves contain more Tic110 protein compared to roots25. These data suggest that the expression of TOC and TIC genes are somehow up-regulated in green tissues, thereby optimizing the accumulation of TOC and TIC proteins where photosynthetic activity is highest. However, whether this regulation is due to photoreceptor-mediated events or developmental regulation remains elusive.
In this study, we investigated the mechanism by which the expression of PhANGs is coordinated with the status of the TOC–TIC complex in Arabidopsis. We demonstrate that the ppi2 mutant possesses active phytochromes and cryptochromes even though the mutant failed to induce the expression of the LHCB gene in response to light. In contrast, many TOC and TIC genes are rapidly induced by blue light in WT and ppi2-2, suggesting that TOC and TIC genes are not controlled by plastid retrograde signals. Furthermore, we also show that light induction of TOC and TIC transcripts is mediated by cryptochrome 1 (CRY1). Based on these results, we discuss the mechanism by which light signals coordinate the expression of PhANGs with plastid protein transport.
Results
Phytochromes and cryptochromes are functional in the ppi2-2 mutant
Previously, we showed that light induction of PhANGs is impaired in the ppi2-2 mutant13. This reduction was most likely due to the action of retrograde signals derived from defective plastids in the ppi2 mutant. However, many PhANGs have been shown to be induced by phytochromes11,12. Therefore, it is also conceivable that the ppi2 mutation reduces the import of enzymes involved in phytochromobilin biosynthesis, thereby affecting the amount of active phytochromes. The reduction of active phytochromes, rather than retrograde signals from defective plastids, might result in the reduced expression of PhANGs in the ppi2-2 mutant.
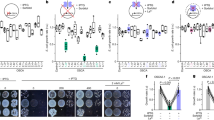
To address this question, we investigated whether the ppi2-2 mutant contains active photoreceptors. Phytochromes have been shown to regulate inhibition of hypocotyl elongation26. Specifically, inhibition of hypocotyl elongation by red-light is mediated by phytochrome B (PHYB), whereas phytochrome A (PHYA) mediates the inhibition of hypocotyl elongation by far-red light27. Therefore, we exposed WT and ppi2-2 plants to continuous red or far-red light for 3 days and measured hypocotyl elongation. As a control, we also investigated hypocotyl elongation of the hy2 mutant that is defective in phytochromobilin biosynthesis16. As shown in Fig. 1, hypocotyl elongation of WT was inhibited by both red and far-red light compared to the hy2 mutant. Far-red light completely inhibited the hypocotyl elongation of the ppi2-2 mutant, indicating that the amount of PHYA in ppi2-2 is comparable to that of WT (Fig. 1B). Continuous red-light did not inhibit the hypocotyl elongation of ppi2-2 completely (Fig. 1A). However, ppi2-2 as well as WT exhibited cotyledon opening upon continuous red-light illumination (Fig. 1A, inlet). In contrast, hy2 failed to exhibit cotyledon opening induced by red light. PHYB has been shown to participate in cotyledon opening induced by red light28,29. Hypocotyl elongation of ppi2-2 was comparable to that of WT in the dark (Supplementary Fig. S1). Hence, we conclude that ppi2-2 mutant contains active PHYB sufficient to induce the red light high irradiance response.
Response of hypocotyl elongation upon monochromatic light irradiation. Plants were grown in the dark for 4 days and then exposed to monochromatic red (A), far-red (B) or blue (C) light for 3 days. Inset in panel (A) shows the magnified images of cotyledons in each genotype. Lower panels show quantitative measurement of hypocotyl elongation during 3-day exposure to red (A), far-red (B) or blue (C) light. Error bars indicate the standard error of the mean (n ≥ 13). Different letters indicate statistically significant differences between genotypes by Tukey–Kramer multiple comparison test (P < 0.05). Bars approximately 1 cm.
We also investigated whether cryptochromes are functional in ppi2-2. CRY1 has been shown to regulate inhibition of hypocotyl elongation by blue light30, and cryptochrome 2 (CRY2) has an additive role in this regulation31. In contrast to the elongated hypocotyl of cry1-500 plants, hypocotyl elongation of the ppi2-2 mutant was completely inhibited by blue light (Fig. 1C). Therefore, we conclude that cryptochromes are also functional in the ppi2-2 mutant.
Taken together, we concluded that PHYA, PHYB and cryptochromes are functional in the ppi2-2 mutant. Given the previously observed down-regulation of PhANGs in ppi2-2, these results also support the hypothesis that plastid signals play key roles in suppressing the expression of PhANGs upon light exposure in ppi2-2.
Blue light induces the expression of TOC and TIC genes
A majority of PhANGs are rapidly induced when etiolated plants are exposed to light. Hence, TOC–TIC complexes must mediate the import of a large number of photosynthesis-associated proteins upon light illumination. However, light-dependent regulation of the TOC–TIC complex has not been analyzed in detail. Therefore, we performed a time-course analysis of TOC and TIC gene expression upon monochromatic light illumination. Both red light and blue light strongly induced the expression of LHCB3.1 in WT (Fig. 2A). In contrast, none of these light treatments induced the expression of LHCB3.1 in the ppi2-2 mutant (Fig. 2B), suggesting that signals derived from ppi2 plastids suppress light-induced PhANG expression. We next investigated the expression of TOC and TIC genes. We found that TOC33, TOC34, TOC75, TOC132, TOC159 and TIC110 genes were induced by blue light in WT (Fig. 2). An intriguing observation was that most of those genes were also induced in the ppi2-2 mutant in response to blue light (Fig. 2). However, this is in contrast to LHCB3.1, which showed a loss of blue-light induction in the ppi2-2 mutant relative to WT. Furthermore, blue light induction of TOC132, a member of the TOC159 family, was stronger in the ppi2-2 mutant than in WT. This was most likely due to compensation for the lack of the TOC159 gene in this mutant.
Time course expression analysis of TOC and TIC genes upon monochromatic light illumination. Plants were grown in the dark for 4 days and then exposed to monochromatic red, far-red or blue light for 0, 2, 4, 8 and 24 h. The mRNA levels were analyzed by real-time PCR and the expression levels were normalized to that of ACTIN2. The expression level of each gene in WT after 24-h red light exposure was set to 1. Error bars represent standard error (SE) of the mean (n = 3).
These data indicate that the expression of TOC and TIC genes are induced by blue light photoreceptors upon light illumination. Furthermore, this induction is not suppressed by retrograde signals derived from ppi2 plastids, as is the case for PhANGs.
Blue light induction of TOC and TIC genes is regulated by cryptochrome 1
The fact that blue light induced the expression of TOC and TIC genes prompted us to further pursue the hypothesis that blue light photoreceptors are responsible for this induction. To this end, we examined whether deficiency of CRY1 affected the blue light induction of TOC and TIC genes using cry1 mutants. As positive controls of CRY1 regulated genes, the expression of CHALCONE SYNTHASE (CHS) and SIGMA FACTOR5 (SIG5) genes was examined32,33. We also examined the expression of ARGININE AMIDOHYDROLASE2 (ARGAH2) that is regulated by CRY234. When etiolated WT plants were exposed to blue light for 8 h, all of the TOC and TIC genes we examined were strongly induced, as were the known CRY1-induced genes, confirming that TOC and TIC are indeed blue light-induced genes (Fig. 3). The blue light induction of CHS and SIG5 was strongly impaired in cry1 mutants, as expected (Fig. 3). The cry1 mutants also failed to induce the expression of TOC and TIC genes examined, with only the exception of TOC34 in cry1-500 mutant (Fig. 3). In contrast, the blue light induction of LHCB3.1 was virtually unaffected by cry1 mutations (Fig. 3). Likewise, the fold change of ARGAH2 by blue light was unaffected or even higher in cry1 mutants (Fig. 3).
Response of TOC and TIC genes to blue light in WT and cry1 mutants. (A) Plants were grown in the dark for 4 days and then exposed to monochromatic blue light for 8 h. CHS and SIG5 genes were selected as the control for CRY1 regulated genes, and ARGAH2 gene was selected as the control for CRY2 regulated gene. The mRNA levels were analyzed by real-time PCR and the expression levels were normalized to that of ACTIN2. The expression level of each gene in WT after 8-h blue light exposure was set to 1. Error bars represent standard error (SE) of the mean (n = 3). (B) Fold change of each gene upon blue light irradiation. The transcript level of each genotype after blue light exposure for 8 h (8 h) was divided by that grown in the dark (0 h).
These data indicate that CRY1 primarily mediates the induction of TOC and TIC genes upon blue light illumination. Because TOC and TIC genes are weakly induced in cry1 mutants, we do not exclude the possibility that other photoreceptors, such as CRY2, participate in the blue light induction of TOC and TIC genes. Nonetheless, the residual induction of TOC and TIC genes in the cry1 mutants was comparable to that of the CHS and SIG5 controls.
Deficiency of cryptochrome 1 affects the accumulation of TOC and TIC proteins
To investigate if a deficiency of CRY1 affects the accumulation of TOC and TIC proteins, we investigated the level of each TOC and TIC protein using immunoblotting. The level of TOC and TIC proteins in the cry1 mutants was comparable to that in WT in the dark (Fig. 4A,B). When the dark grown WT and cry1 plants were subsequently exposed to blue light for 3 days, WT accumulated more LHCP, Toc33, Toc34 and Toc159 proteins compared to cry1 mutants (Fig. 4C). Although the cry1-500 mutant exhibited a stronger phenotype in terms of protein accumulation, results obtained from two independent alleles were consistent. In contrast, Toc75 and Tic110 proteins were virtually unaffected (Fig. 4C). These conclusions were further supported by quantitative analysis of immunoblot signals (Fig. 4D).
Accumulation of TOC and TIC proteins upon exposure to blue light. Plants were grown in the dark for 4 days (A) and then exposed to monochromatic blue light for 3 days (C). Extracted proteins were then resolved by SDS-PAGE, and proteins were probed with antibodies indicated at the left. Protein levels in (A) and (C) were quantified using image acquisition software, normalized to actin levels and shown in (B) and (D), respectively. The level of each protein in WT was set to 1.
In conclusion, deficiency of CRY1 affects the accumulation of TOC and TIC proteins under blue light. This also supports the hypothesis that blue light induction of TOC and TIC genes is a physiologically relevant mechanism to regulate the amount of TOC and TIC proteins.
Discussion
The majority of PhANGs are induced upon light illumination11,12. Hence, plastids must optimize the status of the protein import apparatus, designated as the TOC–TIC complex, in response to light to enable the import of abundant photosynthesis-associated proteins. However, the mechanisms that coordinate the constituents of TOC–TIC complexes with the expression of PhANGs remains to be characterized in detail. In this study, we showed that many of the TOC and TIC genes were induced when etiolated plants were exposed to blue light (Fig. 2). This induction of TOC and TIC genes was largely mediated by CRY1 (Fig. 3). Furthermore, the prolonged blue light exposure affected the accumulation of some TOC proteins in vivo (Fig. 4). Overall, our results suggest that CRY1 is involved in optimizing the status of TOC–TIC complexes in response to abundant precursors of photosynthesis-associated proteins during photomorphogenesis.
Inhibition of hypocotyl elongation by red and far-red light indicates that the ppi2 mutant contains active PHYA and PHYB. It has been shown that genes involved in the chlorophyll branch pathway of tetrapyrrole biosynthesis were down-regulated in ppi2 mutants14. However, ppi2 seems to produce a sufficient amount of phytochrome holoproteins, as illustrated by inhibition of hypocotyl elongation by red and far-red light (Fig. 1). Likewise, hypocotyl elongation was fully inhibited by blue light in ppi2 (Fig. 1), indicating that ppi2 also possesses active cryptochromes. In contrast, ppi2 failed to induce the expression of LHCB through those receptors. These results are consistent with a previous observation that plastid signals can serve as major regulators of light signaling35,36. It is also intriguing that, in mutants with damaged plastids, the light response of PhANGs is much more impaired than that of other phytochrome-regulated genes36. The nature of signals for that regulation is still obscure. However, previous studies showed that PHYTOCHROME-INTERACTING FACTORs (PIFs) and LONG HYPOCOTYL5 (HY5) were involved in the linkage between light signaling and plastid retrograde signaling37,38. Given the fact that both inhibitor treatment and ppi2 mutation exhibited similar effects on light responsiveness of LHCB expression, it is conceivable that common factors are involved in plastid-regulated light responsiveness in both ppi2 and inhibitor-treated plants.
The induction of TOC and TIC genes is mediated by CRY1 rather than phytochromes (Figs. 2, 3). This is in contrast to what we know about the regulation of PhANGs. According to transcriptome analysis, a number of PhANGs are regulated by both phytochromes and cryptochromes19. Thus, light induction of TOC and TIC genes appears to be discriminated from that of PhANGs. The reason why Arabidopsis utilizes CRY1 for the induction of TOC and TIC genes remains obscure. However, it is noteworthy that the induction of LHCB3.1 by blue light is faster than that by red or far-red light (Fig. 2A). In Arabidopsis, the SIG1 gene is also induced by both red and blue light. The expression of SIG1 was strongly induced by red light within 120 min, while blue light induction of SIG1 expression was much faster39. Likewise, the transient induction of LHCB1.3 in Arabidopsis by blue light pulse was faster than that by red light pulse, while plants treated with red light exhibited higher expression of LHCB1.3 in later stages (Gao and Kaufman, 1994). Taken together, these data suggest that the expression of TOC and TIC is rapidly induced by cryptochromes prior to the prolonged induction of PhANGs by phytochromes, allowing timely and efficient transport of photosynthesis-associated proteins. Rapid biogenesis of chloroplasts upon light illumination seems to be, in part, attributable to these mechanisms.
TOC proteins have been shown to be regulated by the ubiquitin proteasome system (UPS)40,41,42,43. Toc159, Toc75 and Toc33 are polyubiquitinated by a RING-type E3 ubiquitin ligase, and lack of this regulation delays de-etiolation upon light illumination42. Given the fact that the level of Toc75 was unaffected even though TOC75 was reduced in the cry1 mutants (Fig. 4), it seems that UPS-dependent regulation of Toc75 predominates over its transcriptional regulation during photomorphogenesis. In contrast, CRY1-dependent transcriptional regulation of TOC159, TOC33 and TOC34 appears to play roles in regulating Toc159, Toc33 and Toc34 protein levels. These data are consistent with the fact that Toc159 and Toc33 families serve as receptor components for precursors2,3,4, and so their levels must be coordinated with light induction of PhANGs. In contrast, Toc75 is involved in the insertion of outer envelope membrane proteins as well as importing chloroplast interior proteins44. As such, levels of TOC and TIC proteins are regulated at multiple levels, allowing plants to efficiently import abundant precursor proteins.
In summary, we have uncovered that TOC and TIC genes, which encode components of the chloroplast protein import apparatus, are induced by blue light through the photoreceptor CRY1. The fact that many photosynthesis-associated proteins, which are the substrates for the TOC–TIC apparatus, also accumulate in response to light illumination suggests that blue light induction of TOC and TIC genes is a part of a mechanism that coordinates PhANG expression with plastid protein import during photomorphogenesis.
Methods
Plant material and growth conditions
All experiments were performed using Arabidopsis thaliana Accession Col-0. The ppi2-2, hy2-101 (a kind gift from Prof. Takayuki Kohchi) and cry1-500 (SALK_042397C; designated as cry1 in a previous report) mutants were described elsewhere13,14,16,45. The cry1-501 (CS303609) and cry1-500 (SALK_042397C) were obtained from Arabidopsis Biological Resource Center and the homozygous T-DNA insertion line was screened by PCR. The homozygous ppi2-2 seeds were obtained using the method as described previously13. Seeds of Arabidopsis thaliana were sterilized with 70% ethanol and 30% bleach solution and then sown on agar plates containing 0.5 × Murashige–Skoog salt and 1% sucrose. To synchronize germination, all seeds were kept at 4 °C for 3 days in the dark.
Monochromatic light sources
Light-emitting diodes (LEDs) were used as the monochromatic light sources. LEDs used in the experiments were as follows (EYELA, Tokyo): Red, STICK-mR LED (λmax = 660 nm at 30 μmol m−2 s−1); Far-red, STICK-mFR (λmax = 735 nm at 25 μmol m−2 s−1); Blue, STICK-mB LED (λmax = 470 nm at 25 μmol m−2 s−1). Unless specified, those light sources were used for monochromatic light irradiation.
Measurement of hypocotyl elongation under monochromatic light irradiation
WT, ppi2, hy2 and cry1 seeds were sown on 0.5 × MS medium containing 1.5% Agar. Prior to dark treatment, seeds on MS plates were irradiated with continuous white light for 8 h at 22 °C. Then, plates were placed vertically and kept in the dark for 4 days at 22 °C. Before monochromatic light treatment, the position of the top of each hypocotyl was marked on plates. Etiolated plants were then irradiated with red light (660 nm), far-red light (735 nm) or blue light (470 nm) at room temperature for 3 days. After the light treatment, hypocotyl elongation during monochromatic light treatment was measured.
Time course analysis of gene expression under monochromatic light
WT and ppi2 mutant seeds were sown on 0.5 × MS medium containing 0.5% Agar. Prior to dark treatment, seeds on MS plates were irradiated with continuous white light at 22 °C for 8 h. Then, plates were kept in the dark for 4 days at 22 °C. Some plants were harvested before monochromatic light treatment. The remaining etiolated plants were then irradiated with red light, far-red light or blue light at room temperature for the times indicated in Fig. 2, and aerial tissues were harvested. The harvested plants were immediately frozen in liquid nitrogen and stored at − 80 °C.
For analysis of TOC and TIC expression in cry1 mutants (Fig. 3), WT and cry1 plants were exposed to blue light for 8 h. Other procedures are the same as stated above.
RNA isolation and real-time PCR analysis
Total RNA was extracted from aerial tissues of wild-type and mutants using RNAiso reagent (Takara). Then, cDNA was synthesized using the PrimeScript reverse transcription (RT) reagent kit (TaKaRa) with random hexamer and oligo(dT) primers. Real-time PCR was performed on a Thermal Cycler Dice Real-Time System TP870 (TaKaRa) using TB Green Premix ExTaq II (TaKaRa) as described previously14,46. Primers used for real-time PCR are listed in Supplementary Table S1. The transcript level of each gene was normalized to that of ACTIN2.
Analysis of TOC and TIC proteins in cry1 mutants
WT and cry1 seeds were sown on MS medium containing 0.5% Agar. Prior to dark treatment, seeds on MS plates were irradiated with continuous white light at 22 °C for 8 h. Plates were kept in the dark for 4 days at 22 °C. Then etiolated plants were exposed to blue light for 3 days at room temperature. After light exposure, green aerial tissues were harvested and frozen in liquid nitrogen, and stored at − 80 °C. Total protein extracts from Arabidopsis were obtained by directly homogenizing leaves in SDS-PAGE sample buffer, as described previously25.
After protein extraction and quantification, the total protein (20 μg or 10 μg) was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using 8%, 12% or 5–20% polyacrylamide gel and immunoblotted with the antisera indicated in the figures. The antibodies against Toc33, Toc34 and Toc159 were kind gifts from Prof. Danny J. Schnell20,47. Tic110 and Toc75 have been previously described25,48. The LHCP antibodies were a kind gift from Prof. Kenneth Cline. Monoclonal antibody against actin was purchased from CHEMICON. Signals were detected using horseradish peroxidase-conjugated secondary antibodies and chemiluminescence reagent. Signals were quantified using image acquisition software (CS Analyzer; ATTO). All the uncropped blots are shown in Supplementary Figs. S2 and S3.
References
Dyall, S. D., Brown, M. T. & Johnson, P. J. Ancient invasions: From endosymbionts to organelles. Science 304, 253–257 (2004).
Richardson, L. G. L. & Schnell, D. J. Origins, function, and regulation of the TOC–TIC general protein import machinery of plastids. J. Exp. Bot. 71, 1226–1238. https://doi.org/10.1093/jxb/erz517 (2020).
Jarvis, P. & Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. https://doi.org/10.1038/nrm3702 (2013).
Li, H. M. & Chiu, C. C. Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61, 157–180 (2010).
Inaba, T. & Schnell, D. J. Protein trafficking to plastids: One theme, many variations. Biochem. J. 413, 15–28 (2008).
Paila, Y. D. et al. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. Elife https://doi.org/10.7554/eLife.12631 (2016).
Li, H. M., Schnell, D. & Theg, S. M. Protein import motors in chloroplasts: On the role of chaperones. Plant Cell 32, 536–542. https://doi.org/10.1105/tpc.19.00300 (2020).
Nakai, M. Reply: The revised model for chloroplast protein import. Plant Cell 32, 543–546. https://doi.org/10.1105/tpc.19.00821 (2020).
de Vries, J., Sousa, F. L., Bolter, B., Soll, J. & Gould, S. B. YCF1: A green TIC?. Plant Cell 27, 1827–1833. https://doi.org/10.1105/tpc.114.135541 (2015).
Nakai, M. YCF1: A green TIC: Response to the de Vries et al. commentary. Plant Cell 27, 1834–1838. https://doi.org/10.1105/tpc.15.00363 (2015).
Terzaghi, W. B. & Cashmore, A. R. Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474 (1995).
Tobin, E. M. & Silverthorne, J. Light regulation of gene expression in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 36, 569–593 (1985).
Tada, A., Adachi, F., Kakizaki, T. & Inaba, T. Production of viable seeds from the seedling lethal mutant pp i2–2 lacking the atToc159 chloroplast protein import receptor using plastic containers, and characterization of the homozygous mutant progeny. Front. Plant Sci. 5, 243. https://doi.org/10.3389/fpls.2014.00243 (2014).
Kakizaki, T. et al. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353 (2009).
Kakizaki, T., Yazu, F., Nakayama, K., Ito-Inaba, Y. & Inaba, T. Plastid signalling under multiple conditions is accompanied by a common defect in RNA editing in plastids. J. Exp. Bot. 63, 251–260. https://doi.org/10.1093/jxb/err257 (2012).
Kohchi, T. et al. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13, 425–436. https://doi.org/10.1105/tpc.13.2.425 (2001).
Terry, M. J. & Smith, A. G. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4, 14. https://doi.org/10.3389/fpls.2013.00014 (2013).
Tepperman, J. M., Zhu, T., Chang, H. S., Wang, X. & Quail, P. H. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442 (2001).
Ma, L. et al. Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. https://doi.org/10.1105/tpc.010229 (2001).
Ivanova, Y., Smith, M. D., Chen, K. & Schnell, D. J. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392 (2004).
Kubis, S. et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16, 2059–2077 (2004).
Vojta, A. et al. The protein translocon of the plastid envelopes. J. Biol. Chem. 279, 21401–21405 (2004).
Li, H. M. & Teng, Y. S. Transit peptide design and plastid import regulation. Trends Plant Sci. 18, 360–366. https://doi.org/10.1016/j.tplants.2013.04.003 (2013).
Kubis, S. et al. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15, 1859–1871 (2003).
Inaba, T. et al. Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17, 1482–1496 (2005).
Quail, P. H. et al. Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. https://doi.org/10.1126/science.7732376 (1995).
Reed, J. W., Nagatani, A., Elich, T. D., Fagan, M. & Chory, J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. https://doi.org/10.1104/pp.104.4.1139 (1994).
Neff, M. M. & Chory, J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. https://doi.org/10.1104/pp.118.1.27 (1998).
Shi, H. et al. The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39, 597–610. https://doi.org/10.1016/j.devcel.2016.10.020 (2016).
Ahmad, M. & Cashmore, A. R. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. https://doi.org/10.1038/366162a0 (1993).
Mazzella, M. A., Cerdan, P. D., Staneloni, R. J. & Casal, J. J. Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128, 2291–2299 (2001).
Moller, S. G., Kim, Y. S., Kunkel, T. & Chua, N. H. PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15, 1111–1119. https://doi.org/10.1105/tpc.008649 (2003).
Fuglevand, G., Jackson, J. A. & Jenkins, G. I. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell 8, 2347–2357. https://doi.org/10.1105/tpc.8.12.2347 (1996).
Ohgishi, M., Saji, K., Okada, K. & Sakai, T. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101, 2223–2228. https://doi.org/10.1073/pnas.0305984101 (2004).
Ruckle, M. E., Burgoon, L. D., Lawrence, L. A., Sinkler, C. A. & Larkin, R. M. Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol. 159, 366–390. https://doi.org/10.1104/pp.112.193599 (2012).
Vinti, G., Fourrier, N., Bowyer, J. R. & Lopez-Juez, E. Arabidopsis cue mutants with defective plastids are impaired primarily in the photocontrol of expression of photosynthesis-associated nuclear genes. Plant Mol. Biol. 57, 343–357. https://doi.org/10.1007/s11103-004-7867-8 (2005).
Martin, G. et al. Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 7, 11431. https://doi.org/10.1038/ncomms11431 (2016).
Ruckle, M. E., DeMarco, S. M. & Larkin, R. M. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960. https://doi.org/10.1105/tpc.107.054312 (2007).
Onda, Y., Yagi, Y., Saito, Y., Takenaka, N. & Toyoshima, Y. Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: Differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J. 55, 968–978. https://doi.org/10.1111/j.1365-313X.2008.03567.x (2008).
Shanmugabalaji, V. et al. Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 28, 2616–2623. https://doi.org/10.1016/j.cub.2018.06.006 (2018).
Thomson, S. M., Pulido, P. & Jarvis, R. P. Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 48, 71–82. https://doi.org/10.1042/BST20190274 (2020).
Ling, Q., Huang, W., Baldwin, A. & Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659. https://doi.org/10.1126/science.1225053 (2012).
Ling, Q. et al. Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science https://doi.org/10.1126/science.aav4467 (2019).
Tu, S. L. et al. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16, 2078–2088 (2004).
Lee, B. R., Koprivova, A. & Kopriva, S. The key enzyme of sulfate assimilation, adenosine 5’-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. Plant J. 67, 1042–1054. https://doi.org/10.1111/j.1365-313X.2011.04656.x (2011).
Tokumaru, M. et al. Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol. 173, 524–535. https://doi.org/10.1104/pp.16.01546 (2017).
Smith, M. D. et al. atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165, 323–334 (2004).
Okawa, K. et al. Targeting of a polytopic membrane protein to the inner envelope membrane of chloroplasts in vivo involves multiple transmembrane segments. J. Exp. Bot. 65, 5257–5265. https://doi.org/10.1093/jxb/eru290 (2014).
Acknowledgements
We thank Ms. Saori Hamada and Fumi Adachi (University of Miyazaki) for their technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (18H02169 to T. I. and 20H02917 to Y. I. I.), the Public Foundation of Elizabeth Arnold-Fuji, and a Grant for Scientific Research on Priority Areas from the University of Miyazaki (to T.I.). L.G.L.R. was supported by a JSPS-NSERC Summer Program Fellowship. S.U. was supported by a JSPS Fellowship for Young Scientists.
Author information
Authors and Affiliations
Contributions
L.G.L.R., T.K., Y.I.I. and T.I. contributed to the experimental design. T.K., Y.I.I. and T.I. supervised the research. H.F., A.T., L.G.L.R., T.K., S.U. and T.I. performed the experiments. All authors analyzed the data. T.I. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fukazawa, H., Tada, A., Richardson, L.G.L. et al. Induction of TOC and TIC genes during photomorphogenesis is mediated primarily by cryptochrome 1 in Arabidopsis. Sci Rep 10, 20255 (2020). https://doi.org/10.1038/s41598-020-76939-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-76939-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.