Abstract
Jagged1 activates Notch signaling and subsequently promotes osteogenic differentiation in human periodontal ligament cells (hPDLs). The present study investigated the participation of the Notch receptor, NOTCH2, in the Jagged1-induced osteogenic differentiation in hPDLs. NOTCH2 and NOTCH4 mRNA expression levels increased during hPDL osteogenic differentiation. However, the endogenous NOTCH2 expression levels were markedly higher compared with NOTCH4. NOTCH2 expression knockdown using shRNA in hPDLs did not dramatically alter their proliferation or osteogenic differentiation compared with the shRNA control. After seeding on Jagged1-immobilized surfaces and maintaining the hPDLs in osteogenic medium, HES1 and HEY1 mRNA levels were markedly reduced in the shNOTCH2-transduced cells compared with the shControl group. Further, shNOTCH2-transduced cells exhibited less alkaline phosphatase enzymatic activity and in vitro mineralization than the shControl cells when exposed to Jagged1. MSX2 and COL1A1 mRNA expression after Jagged1 activation were reduced in shNOTCH2-transduced cells. Endogenous Notch signaling inhibition using a γ-secretase inhibitor (DAPT) attenuated mineralization in hPDLs. DAPT treatment significantly promoted TWIST1, but decreased ALP, mRNA expression, compared with the control. In conclusion, Notch signaling is involved in hPDL osteogenic differentiation. Moreover, NOTCH2 participates in the mechanism by which Jagged1 induced osteogenic differentiation in hPDLs.
Similar content being viewed by others
Introduction
Notch signaling regulates various cell responses during development and homeostasis, including cell proliferation, survival, and differentiation. The binding of a transmembrane Notch ligand to a Notch receptor on adjacent cells activates intracellular Notch signaling by releasing the Notch intracellular domain (NICD) of the receptor. The NICD translocates into the nucleus and binds to a transcriptional complex, turning this transcriptional repressor to an activator complex. The mammalian canonical Notch receptors and ligands are NOTCH1, NOTCH2, NOTCH3, NOTCH4, and DLL1, DLL3, DLL4, JAG1, and JAG2, respectively1,2.
The role of Notch signaling in dental-related cells has been reported. Notch signaling directly and indirectly regulates osteoclastogenesis. In orthodontic-treated rodent teeth, Notch receptors and ligands are upregulated in bone resorptive sites3. Jagged1 decreases osteoprotegerin (OPG), but not receptor activator of nuclear factor-κB ligand (RANKL), expression in human periodontal ligament cells (hPDLs)4. Hence, the OPG/RANKL ratio is reduced, favoring osteoclast formation. Correspondingly, Jagged1 directly influences osteoclast differentiation in a murine macrophage-like cell line5. Moreover, Notch signaling increases PDL cell viability under stress6. Jagged1 immobilization activates Notch signaling and promotes osteogenic differentiation in hPDLs7,8. Jagged1-treated hPDLs exhibit increased alkaline phosphatase (ALP) enzymatic activity and an in vitro mineral deposition compared with cells in the control condition. Furthermore, osteogenic medium upregulates the mRNA expression of the Notch ligand, DLL1, in hPDLs9. Adding a Notch signaling inhibitor attenuates osteoblast maturation as determined by decreased mineral deposition10. Taken together, these data suggest the role of Notch signaling in hPDL homeostasis.
A previous study demonstrated that hPDLs express all 4 Notch receptors, NOTCH1-44. The extracellular domain of the Notch receptor consists of epidermal growth factor (EGF)-like repeats that function as ligand binding sites. Different Notch receptors contain different numbers of repeats and exhibit different ligand binding properties and preferentially bind with certain ligands11. It has been reported that each Notch receptor influences different cell responses11. Notch receptors are expressed in specific locations during organ development and mutation of each receptor leads to dissimilar phenotypes11,12,13. Hence, the specific roles of each Notch receptor should be determined. The present study investigated the participation of NOTCH2 in Jagged-induced osteogenic differentiation in hPDLs.
Results
Notch receptors and ligands are expressed during hPDL osteogenic differentiation
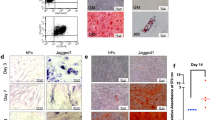
hPDLs differentiated into the osteogenic lineage when the cells were incubated in osteogenic medium (OM). A significant increase in mineral deposition was observed at 14 and 21 days in OM (Fig. 1A,B). The mRNA expression levels of Notch receptors, ligands, and target genes were examined using real-time PCR at day 0, day 5, day 10, day 14, and day 21 during osteogenic differentiation. Among the Notch receptors, NOTCH3 mRNA levels did not change during the observation period. The increased expression of NOTCH2 and NOTCH4 was observed during osteogenic differentiation while NOTCH1 mRNA levels significantly decreased at day 21 compared with day 0 (Fig. 1C–F). A significant increase from day 0 was noted at day 10 for NOTCH2, and at day 5 and day 10 for NOTCH4. A significant upregulation of Notch ligands (JAG1 and DLL1) was detected at day 5 and day 10 for JAG1 and at day 5, day 10, and day 14 for DLL1 during osteogenic differentiation compared with day 0 (Fig. 1G–L). DLL3 mRNA levels significantly decreased at day 14 and day 21 compared with day 0. In contrast, there was no marked difference in JAG2, DLL4, and HES1 mRNA expression during osteogenic differentiation (Fig. 1G–L).
The mRNA expression of Notch receptors, ligands, and target genes during hPDL osteogenic differentiation. hPDLs were maintained in growth medium (GM) or osteogenic medium (OM). Mineral deposition was determined by alizarin red s staining at day 14 and 21 (A). The stained dye was solubilized and measured at an absorbance at 570 nm (B). The mRNA expression of Notch receptors, ligands, and target genes was evaluated by real-time PCR at day 0, day 5, day 10, day 14, and day 21 (C–L). Bars indicate a significant difference.
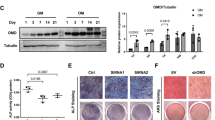
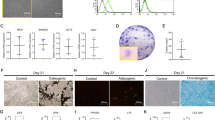
The baseline mRNA expression of Notch receptors was evaluated using real-time PCR. The results demonstrated that hPDLs expressed all four types of Notch receptors, with NOTCH2 mRNA levels being the most highly expressed (Fig. 2A). Correspondingly, protein expression of all four types of Notch receptors was evaluated using flow cytometry analysis (Fig. 2B,C). NOTCH2 protein was the most expressed Notch receptor in hPDLs.
The baseline mRNA expression of the four Notch receptors in hPDLs was examined using real-time PCR (A). Protein expression levels were examined using flow cytometry analysis. The representative histogram was illustrated (B) and the mean fluorescence intensity per cells was demonstrated (C). Cells were transduced with either lentiviral particles containing shRNA scramble sequence (shControl) or shRNA against NOTCH2 (shNOTCH2). The NOTCH2 knockdown efficiency was examined using real-time PCR (D) and flow cytometry analysis (E and F). Bars indicate a significant difference.
NOTCH2 knockdown did not alter cell proliferation or mineralization
Because NOTCH2 was the highest expressed Notch receptor in hPDLs and significantly increased during osteogenic differentiation, hPDLs were subjected to knockdown of NOTCH2 expression using shRNA. The hPDLs were transduced with lentiviral particles containing shRNA against NOTCH2 (shNOTCH2). Lentiviral particles containing a scrambled shRNA sequence were employed in the control condition (shControl). PCR and flow cytometry analysis were performed to confirm the NOTCH2 knockdown. A significant reduction in NOTCH2 mRNA and protein levels was observed in shNOTCH2 cells (Fig. 2D–F). NOTCH2 mRNA and protein expression were 22.38% and 37.4% compared with the control condition, respectively. There was no significant difference in other Notch receptor mRNA levels between the shControl and shNOTCH2 groups (Fig. 3A–C). A significant increase in DLL1, DLL3, and DLL4 mRNA levels was detected (Fig. 3D–F). However, JAG1, JAG2, HES1, and HEY1 mRNA expression were similar between the groups (Fig. 3G–J).
Effect of NOTCH2 knockdown on cell proliferation and osteogenic differentiation in hPDLs. Cells were transduced with either lentiviral particles containing shRNA scramble sequence (shControl) or shRNA against NOTCH2 (shNOTCH2). The mRNA expression of Notch pathway components was examined using real-time PCR (A–J). Cell viability was determined at day 1, 3, and 7 days (K). shControl and shNOTCH2 cells were maintained in either growth medium or osteogenic medium for 21 days and mineral deposition was stained with alizarin red s dye (L). Absorbance values of solubilized dye (M). Osteogenic marker gene expression was evaluated using real-time PCR at day 7 after osteogenic differentiation (N). Bars indicate a significant difference. Asterisks indicate a significant difference compared with shControl.
Both shControl and shNOTCH2 cells demonstrated a significant increase in cell number at day 7 compared with day 1 (Fig. 3K). However, there was no significant difference between these two groups at the observed time points. When the cells were maintained in OM for 21 days, both shControl and shNOTCH2 cells formed mineral deposits in OM (Fig. 3L). A significant increase in mineral precipitation was present in OM compared with growth medium (GM); however, there was no dramatic change in mineralization in the shNOTCH2 group compared with the shControl group (Fig. 3M). Correspondingly, the ALP and OSX mRNA levels in the cells maintained in OM for 7 days were similar between the shControl and shNOTCH2 groups (Fig. 3N). Although there was a significant difference in RUNX2 and OCN mRNA levels in the shNOTCH2 group compared with the shControl group, the change was less than 0.25-fold.
NOTCH2 participated in Jagged1-induced osteogenic differentiation in hPDLs
Previous reports demonstrated that indirect affinity-immobilized Jagged1 promoted osteogenic differentiation in hPDLs as determined by increased ALP activity, osteogenic marker gene expression, and mineralization7,8,14. The hPDLs seeded on Jagged1 surfaces demonstrated dramatically enhanced an in vitro mineralization compared with those on the hFc control surfaces when the cells were maintained in OM for 14 days (Fig. 4A,B).
shNOTCH2 cells exhibited less ALP enzymatic activity and mineralization in Jagged1 treated condition. hPDLs were seeded on Jagged1-immobilized surfaces. The control condition was hFc-immobilized surfaces. Mineral deposition was examined using alizarin red s staining at day 14 (A). Absorbance of the solubilized dye (B). ALP staining and enzymatic activity assay were evaluated at day 3 and 7 after osteogenic differentiation (C–E). An in vitro mineralization was stained with alizarin red s dye (F). Absorbance of the solubilized dye (G). Bars indicate a significant difference. GM growth medium, OM osteogenic medium.
shControl and shNOTCH2 cells were seeded on Jagged1-immobilized surfaces and cultured in OM. ALP enzymatic activity and osteogenic marker gene expression were evaluated at day 3 and day 7. Mineral deposition was examined using alizarin red s staining, SEM, and EDX at day 14. Jagged1-induced ALP enzymatic activity was markedly increased in the shControl, but not in the shNOTCH2 cells at both day 3 and day 7 (Fig. 4C–E). In addition, Jagged1-induced mineral deposition was attenuated in the shNOTCH2 cells, while the shControl cells on Jagged1 surfaces exhibited a substantial increase in mineralization compared with those on hFc control surfaces (Fig. 4F,G). Jagged1-induced COL1 protein expression was markedly attenuated in shNOTCH2 cells compared with those detected in the shControl cells at both day 3 and day 7 after osteogenic differentiation (Fig. 5).
shNOTCH2 cells expressed less collagen type I in Jagged1 treated condition. hPDLs were seeded on Jagged1-immobilized surfaces. The control condition was hFc-immobilized surfaces. Collagen type I protein expression was examined using immunofluorescence staining at day 3 and day 7 after osteogenic differentiation. White bars indicate 50 μm.
The mineral crystals in Jagged1-treated shNOTCH2 cells were fewer and smaller compared with the Jagged1-treated shControl cells (Fig. 6A). However, the chemical composition of the mineral crystals was similar between the shControl and shNOTCH2 cells (Fig. 6B). The mRNA expression was examined at day 3 and day 7 (Fig. 7). Reduced NOTCH2 mRNA levels in the shNOTCH2 cells were consistently evident compared with the shControl cells in OM (Fig. 7A,I). shNOTCH2 cells exhibited lower Notch target gene, HES1 and HEY1, expression levels after exposure to Jagged1-immobilized surfaces (Fig. 7B,C,J,K). HES1 and HEY1 mRNA levels were markedly upregulated in Jagged1 treated shControl cells, however there was no significant difference in HES1 and HEY1 mRNA expression in Jagged1-treated shNOTCH2 cells compared with the hFc control. MSX2, ALP, and COL1A1 mRNA levels were significantly enhanced in the shControl cells on Jagged1 surfaces compared with those on hFc surfaces (Fig. 7D,F,L,N). A negative regulator of osteogenic differentiation, TWIST2, mRNA levels were significantly decreased in Jagged1-treated shControl cells (Fig. 7G,H,O,P). None of the osteogenic maker genes were significantly altered in shNOTCH2 cells in the Jagged1 condition compared with those in the hFc condition.
shNOTCH2 cells exhibited less mineralization in Jagged1-treated condition. shControl and shNOTCH2 cells were seeded on Jagged1-immobilized surfaces. The control condition was hFc-immobilized surfaces. Mineral crystal morphology and composition were determined using scanning electron microscope (A) and energy-dispersive X-ray spectroscopic analysis (B) at day 14 after osteogenic differentiation.
shNOTCH2 attenuated the effect of Jagged1 on Notch target gene and osteogenic marker gene expression. shControl and shNOTCH2 cells were seeded on Jagged1-immobilized surfaces. The control condition was hFc-immobilized surfaces. The mRNA expression was evaluated using real-time PCR at day 3 and day 7 after osteogenic differentiation. Bars indicate a significant difference.
Notch signaling inhibition attenuated mineralization in hPDLs
To evaluate the role of endogenous Notch signaling in hPDL osteogenic differentiation, cells were maintained in OM with or without a γ-secretase inhibitor, DAPT. DAPT treatment attenuated mineral deposition in hPDLs at 14 and 21 days after osteogenic differentiation (Fig. 8A,B). HES1, but not HEY1, mRNA was significantly decreased in the DAPT-treated condition at the observed time points (Fig. 8C,D). Because DAPT decreased mineralization in hPDLs, the mRNA expression of the phosphate regulatory genes was investigated. DAPT-treatment significantly decreased ALP, but increased PIT1 mRNA levels, at all time points (Fig. 8E,F). ENPP mRNA expression was significantly decreased at day 3, while there was no change in ANKH mRNA levels in the DAPT-treated group (Fig. 8G,H). OPN mRNA expression increased at all time points, however, the difference was signficant only at day 3 (Fig. 8I). For osteogenic-related transcription factors, hPDLs treated with DAPT demonstrated similar RUNX2 mRNA levels compared with the control condition, while markedly reduced MSX2 expression levels were observed at day 3 (Fig. 8J,K). There was no significant change in the mRNA levels of DMP1, DSPP, or OCN in the DAPT-treated group compared with the control (Fig. 8L,M,O) . However, decreased COL1A1 expression levels were noted at day 3 (Fig. 8N). TWIST1, but not TWIST2, mRNA levels were significantly upregulated in the DAPT-treated hPDLs (Fig. 8P,Q).
Endogenous Notch signaling participated in mineralization in hPDLs. hPDLs were maintained in osteogenic medium (OM) with or without a γ-secretase inhibitor (DAPT). Mineralization was determined using alizarin red s staining at day 14 and 21 (A). The absorbance at 570 nm of the solubilized dye (B). The mRNA expression was examined using real-time PCR at day 1, 3, and 7 (C–Q). Bars indicate a significant difference.
Effect of endogenous Notch signaling inhibition on osteogenic related gene expression was also evaluated in the GM. hPDLs were cultured in GM and treated with DAPT for 1, 3, and 7 days. Results demonstrated that DAPT was able to inhibit HES1 mRNA expression, confirming Notch signaling inhibition (Fig. 9A). DAPT treated hPDLs in growth medium significantly decreased ALP mRNA levels (Fig. 9C). Although, there were significant difference of ANKH, DSPP, and TWIST1 mRNA levels between the control and DAPT treated condition. The fold change was less than 1.5 (Fig. 9).
Discussion
The present study described the role of NOTCH2 in immobilized Jagged1-induced osteogenic differentiation in hPDLs. Previous report demonstrated that Jagged1 promoted osteogenic differentiation in hPDLs. In the present study, we described in detail on the receptor participating in this regulation. NOTCH2 mRNA and protein levels is the highest among Notch receptors in hPDLs and significantly increased during osteogenic differentiation. NOTCH2 knockdown resulted in the marked decrease of ALP enzymatic activity, collagen type I expression, and mineral deposition. Hence, the present study highlights the participation of NOTCH2 in mechanism by which Jagged1 induced osteogenic differentiation in hPDLs.
Notch signaling regulates osteogenic differentiation. However, this effect is cell type and stage specific15. The Notch ligand, Jagged1, markedly promoted osteogenic differentiation in hPDLs as determined by a significant increase in ALP enzymatic activity, osteoblast marker gene expression, and mineral deposition in vitro8,16. Corresponding, Jagged1-induced osteogenic differentiation was observed in human mesenchymal stem cells (hMSCs), human dental pulp cells, stem cells isolated from human exfoliated deciduous teeth, and human bone-derived cells7,15,17,18,19,20. In contrast, Jagged1 did not dramatically promote osteogenic differentiation in retinoic acid treated mouse induced pluripotent stem cells21. The mechanism(s) related to this phenomenon remains unclear. It was shown that a PKCδ inhibitor attenuated Jagged1-induced osteogenic differentiation in hMSCs15, implying that PKCδ participates in the regulatory mechanism. A study in vascular smooth muscle cells illustrated that the Notch1 intracellular domain enhanced osteogenic differentiation via increased Msx2 transcriptional activity22. The present study demonstrated the involvement of NOTCH2 in Jagged1-induced MSX2 expression in hPDLs. The shControl cells upregulated MSX2 mRNA expression after exposure to Jagged1, while this phenomenon was attenuated in Jagged1-treated shNOTCH2 cells.
A previous study in human periodontal ligament stem cells (hPDLSCs) showed that Jagged1 promoted osteogenic differentiation by downregulating a negative regulator of osteogenic differentiation. Jagged1-treated hPDLSCs had a marked reduction of TWIST family gene expression8. However, the direct relation of Notch receptors and TWIST promoter activity has not been reported. A Twist/Notch correlation has been demonstrated in Drosophila. Notch activation caused Twist expression23. Drosophilla with reduced Twist or Notch expression levels exhibited similar muscle development phenotypes23. TWIST1 and TWIST2 also function as negative regulators of osteogenic differentiation by directly binding to the RUNX2 promoter, inhibiting RUNX2 transcription24. TWIST knockdown in hPDLs resulted in the increased mRNA expression of osteogenic marker genes and alkaline phosphatase enzymatic activity25. The present study demonstrated that endogenous Notch signaling inhibition resulted in increased TWIST1 mRNA levels in hPDLs. Correspondingly, TWIST2 mRNA expression was attenuated in Jagged1-treated shControl cells, while this effect was rescued in NOTCH2 knockdown cells, confirming that Notch promotes osteogenic differentiation by downregulating TWIST gene expression via Jagged1/Notch2 binding.
hPDLs expressed all 4 types of canonical Notch receptors, NOTCH1-4. NOTCH2 mRNA and protein levels were the highest among the Notch receptors. This result is consistent with previous studies showing high expression of NOTCH2 in hPDL cells4,26. Different types of Notch receptors function differently when binding to the same ligand27,28. NOTCH2 plays a role in hPDL homeostasis, because it has been shown that orthodontic force application in rat molars led to upregulated Notch2 expression in rat periodontal tissues3. NOTCH2 knockdown in hPDLs decreased the effect of Jagged1-induced osteoprotegerin expression4. In addition, NOTCH2 mRNA expression was significantly increased after hPDLs were exposed to an intermittent compressive force for 24 h26. The upregulation of NOTCH2 under intermittent stress also participated in the mechanism by which TGF-β1 regulated sclerostin expression in hPDLs26. However, the role of NOTCH2 in hPDL osteogenic differentiation remains unknown.
The function of Notch2 in osteogenic differentiation and bone formation is unresolved. Conditional knockout of Notch2 driven by Runx2 expression in cells in mice resulted in increased trabecular bone mass, implying that Notch2 inhibits trabecular bone formation29. In contrast, a time course study in an MG63 osteosarcoma cell line found that NOTCH1 and NOTCH3 expression decreased in the early phase and NOTCH2 and NOTCH4 expression increased in the later phase during osteogenic differentiation in vitro30. Correspondingly, the present study in hPDLs observed a slight decrease in NOTCH1 expression during the osteogenic differentiation time course, while NOTCH2 and NOTCH4 mRNA levels were upregulated. NOTCH1, NOTCH2, and NOTCH4 mRNA levels were upregulated in human alveolar bone-derived osteoprogenitor cells seeded on modified titanium surfaces that correlated to increased osteogenic marker gene expression, implying the involvement of these receptors in osteogenic differentiation31. Previous research using human dental pulp cells illustrated that HES1 expression increased during odonto/osteogenic differentiation19. The present study also observed a slight increase in HES1 mRNA levels during hPDL osteogenic differentiation; however, the increase was not significant. This evidence suggests that Notch signaling positive regulates hPDL osteogenic differentiation.
shNotch2 cells exhibited the impairment of Jagged1-induced osteogenic differentiation. However, NOTCH2 knockdown did not markedly affect osteogenic differentiation in hPDLs in condition without immobilized Jagged1, despite the fact that hPDLs expressed JAG1. As mentioned earlier, Notch signaling regulating cell response is complex and context dependent. Several hypotheses are postulated to address this point. First, we suspect that endogenous Notch signaling is dispensable during osteogenic differentiation process in hPDLs. Despite the fact that endogenous Notch signaling inhibition attenuated mineralization, DAPT treatment did not dramatically alter the expression levels of key osteogenic related transcriptional factors, RUNX2 and MSX2. Although, the significant increase of TWIST1 mRNA levels was observed. The fold change was less than 1.5. Second, hPDLs expressed both NOTCH2 and Jagged1. We hypothesize that endogenous Jagged1 protein expression levels are low, unlike the abundant Jagged1 immobilization on tissue culture surface. As Notch signaling initiates by the binding of transmembrane Notch receptor and ligand. There is no intracellular signaling amplification. Hence, the endogenous expression levels of both Notch receptors and ligands are indeed crucially impact on signal transduction and subsequently cell responses. Further study is necessitated to determine the endogenous protein levels of Jagged1 in hPDLs. Third, shNOTCH2 cells exhibited a slight change in Notch expression phenotypes. As shown in Fig. 3, shNOTCH2 cells significantly increased the expression of DLL ligands which could bind to other receptors and compensate the effect on osteogenic differentiation in shNOTCH2.
Inhibiting endogenous Notch signaling with a γ-secretase inhibitor (DAPT) resulted in attenuated mineral deposition without a marked change in osteogenic marker gene expression. It is hypothesized that endogenous Notch signaling controls phosphate/pyrophosphate metabolism and subsequently influence mineralization. DAPT suppressed ALP expression, while the ANKH mRNA levels did not change. This could result in a decreased phosphate/pyrophosphate ratio, which inhibits mineralization. In addition, OPN mRNA levels were increased in DAPT-treated hPDLs. OPN is an extracellular matrix component and considered an osteogenic differentiation marker. OPN is a potent mineralization inhibitor32. OPN binds to mineral crystals and prevents their growth32. Taken together, the potential reduced Pi/PPi ratio and increased OPN expression participate in the DAPT-attenuated mineralization in hPDLs.
In summary, the present study illustrated that NOTCH2 participates in Jagged1-induced osteogenic differentiation in hPDLs because shNOTCH2-transduced cells exhibited the attenuated Jagged1-induced ALP enzymatic activity, osteogenic related gene expression, and mineral deposition compared with shControl cells. Further, direct evidence of the NOTCH2 intracellular domain interaction with the osteogenic related gene promotors is required to confirm the direct influence of NOTCH2.
Materials and methods
Cell isolation and culture
All experimental protocols and the study design was approved by the Human Research Ethics Committee, Faculty of Dentistry, Chulalongkorn University (approval no. 089/2015). All experimental protocols were carried out in accordance with the Helsinki declaration and relevant guideline and regulations. Informed consent was obtained. Embedded and impacted third molars without pathological conditions scheduled for surgical removal were collected for cell isolation. Periodontal ligament tissues were gently scraped from the middle 1/3 of the root to prevent gingival and cellular cementum cell contamination. The tissues were minced into small pieces and placed on 35-mm tissue culture dishes for cell explantation. The cells were maintained in growth medium (GM) composed of Dulbecco’s modified Eagle medium (DMEM, cat. no. 11960, Gibco, USA) containing 10% fetal bovine serum (FBS, cat. no. 10270, Gibco, USA), 2 mM L-glutamine (GlutaMAX-1, cat. no. 35050, Gibco, USA), 100 unit/mL penicillin, 100 μg/mL streptomycin and 250 ηg/mL amphotericin B (Antibiotic–Antimycotic, cat. no. 15240, Gibco, USA). The cells were incubated at 37 °C in a humidified 5% carbon dioxide atmosphere. The culture medium was changed every 48 h. The cells were sub-cultured at a 1:3 ratio when confluent. The experiments employed cells from passages 3‒6.
Lentiviral shRNA transduction
Lentiviral shRNA transduction was performed in hPDLs with human NOTCH2 shRNA lentiviral particles (cat. no. sc-40135-v; Santa Cruz Biotechnology, Dallas, TX, USA) or the control viral particle (cat. no. sc-108080; Santa Cruz Biotechnology). The transduction processes were performed according to the manufacturer’s protocol. Briefly, hPDLs were incubated in lentivirus transduction solution (5 × 104 infectious units of virus) with 10 μg/mL polybrene (cat. no. sc-134220, Santa Cruz Biotechnology) for 18 h. The transduced hPDLs were selected with 10 μg/mL puromycin (cat. no. 108071, Santa Cruz Biotechnology).
Osteogenic differentiation
hPDLs were seeded at a density of 75,000 cells/well on 24-well-plates and maintained in growth medium for 24 h. Subsequently, the culture medium was changed to osteogenic medium (OM), which was growth medium supplemented with 50 μg/mL ascorbic acid (cat. no. A-4034, Sigma-Aldrich, St. Louis, MO, USA), 250 nM dexamethasone (cat. no. D8893, Sigma-Aldrich), and 5 mM β-glycerophosphate (cat. no. G9422, Sigma-Aldrich). In some experiments, the osteogenic medium was supplemented with a γ-secretase inhibitor, DAPT (20 μM, cat. no. D5942, Sigma-Aldrich).
Jagged1 treatment
Jagged1 was immobilized on the tissue culture surface using a previously described indirect affinity immobilization method8. Briefly, the tissue culture surfaces were incubated with recombinant protein G (50 μg/mL, cat. no. 101201, Invitrogen, Rockford, IL, USA) for 16 h, followed by coating the surface with bovine serum albumin solution (10 mg/mL, cat. no. A9418, Sigma-Aldrich) for 2 h. The tissue culture surfaces were rinsed with sterile phosphate buffered saline between steps. Lastly, the surfaces were exposed to Jagged1/Fc (10 nM, cat. no. 1277-JG, R&D Systems, Minneapolis, MN, USA) for 2 h. Human IgG Fc fragments (hFc, cat. no. 009000008, Jackson Immuno Research Labs, USA) were used in the control conditions.
Polymerase chain reaction (PCR)
Total cellular RNA was isolated using Trizol reagent (RiboEx solution, cat. no. 301-001, GeneAll, Seoul, South Korea). Total RNA (1 μg) was converted to cDNA using reverse transcriptase (ImProm-II Reverse Transcription System, cat. no. A3800, Promega, Madison, WI, USA). Real-time PCR was performed in a CFX connect Real-Time PCR (Bio-Rad, Singapore) with FastStart Essential DNA Green Master (Roche Diagnostic, Mannheim, Germany). The real-time PCR reaction started with denaturing at 95 °C for 5 min, followed by 40 cycles of amplification, and ended with an extension reaction at 72 °C for 20 min. The amplification cycles were (1) 95 °C for 20 s, (2) 60 °C for 20 s, and (3) 72 °C for 20 s. Melt curve analysis was performed to determine product specificity. Relative gene expression was calculated using the 2−ΔΔCt method33. The reaction product was quantified with GAPDH as the reference gene. For endpoint PCR, the reaction was performed with Taq polymerase (Roche Applied Science, IN, USA) in a thermocycler machine. The amplified products were electrophoresed in 1.8% agarose gel and stained with ethidium bromide. The endpoint PCR reaction started with denaturing at 95 °C for 2 min, followed by 35 cycles of amplification, and ended with an extension reaction at 72 °C for 7 min. The amplification cycles were (1) 94 °C for 45 s, (2) 60 °C for 60 s, and (3) 72 °C for 90 s. Band density was evaluated using ImageJ software. The oligonucleotide sequences used are shown in Suppl Table 1.
MTT assay
hPDLs were incubated with 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyltetrazolium bromide solution (cat. no. M6494, Invitrogen) at a final concentration of 0.5 mg/ml. The cells were maintained at 37 °C for 30 min to allow formazan crystal formation. Subsequently, the precipitated formazan was solubilized in dimethylsulfoxide/glycine buffer. The solution was measured an absorbance at 570 nm.
ALP activity and staining
Cells were lysed with alkaline lysis buffered solution. An aliquot of the solution was subjected to ALP activity assay and total protein assay. For the ALP activity assay, an aliquot of the cell lysate was incubated with p-nitrophenol phosphate solution (PNPP, cat. no. 002201, Life technologies, Frederick, MD, USA) at 37 °C for 15 min. The reaction was stopped by adding 0.1 M NaOH. The products were measured at an absorbance of 410 nm. For total protein, an aliquot of the cell lysate was incubated with bicinchoninic acid solution (Pierce BCA protein assay kit, ThermoFisher Scientific, USA) and the assay was performed according to the manufacturer’s protocol. For ALP staining, the cells were fixed with 10% buffered formalin for 10 min. After rinsing with phosphate buffered saline, the cells were stained with BCIP/NBT tablets (Roche, USA) for 30 min and kept out from light at room temperature.
Scanning electron microscopic and energy-dispersive X-ray spectroscopic analysis
Cells were fixed with 2.5% glutaraldehyde solution (cat. no. 49629, Sigma-Aldrich) for 10 min and dehydrated with a graded series of ethanol solutions. The samples were processed for critical point drying. The cell and mineral morphology were observed using a scanning electron microscope (SEM; Quanta 250, FEI, Netherlands). The chemical composition was determined using energy-dispersive X-ray spectroscopic analysis (EDX).
Alizarin red s staining
Samples were fixed with cold methanol for 10 min, washed with deionized water, and stained with a 2% alizarin red s solution for 3 min at room temperature. The stained dye was solubilized with 10% cetylpyridinium chloride monohydrate in 10 mM sodium phosphate at room temperature with gently agitation for 15 min. The solubilized dye was measured an absorbance of 570 nm.
Flow cytometry analysis
Cells were harvested to create single cell suspension and further incubated with fluorochrome labelled antibodies (dilution 1:5) for 30 min at 4 °C in PBS containing 1% FBS. The stained cells were analysed on FACSCelesta instrument (BD Bioscience). Data were presented as mean fluorescence intensity per cell. The antibodies were BB515-conjugated mouse anti-human NOTCH1 (clone MHN1-519; BD Horizon), PE-conjugated mouse anti-human NOTCH2 (clone MHN2-25; BD Pharmingen), PE-conjugated mouse anti-human NOTCH3 (clone MHN3-21; BD Pharmingen), and PE-conjugated mouse anti-human NOTCH4 (clone MHN4-2; BD Pharmingen). Fluorescence-conjugated mouse IgG was employed as the isotype control.
Immunofluorescense staining
Cells were fixed with 4% buffered formalin for 10 min. Horse serum (2% v/v) was used to inhibit a non-specific binding. Cells were stained with mouse anti-collagen I (C2456, Sigma) at 4 °C overnight and subsequently incubated with biotinylated anti-mouse antibodies (Invitrogen) for 40 min. The targeted protein expression was visualized by stained with Strep-FITC (Sigma). DAPI (TOCRIS bioscience) was employed for nuclei staining.
Statistical analyses
The data were presented as mean ± standard error of the mean. Each dot represents each data point. Statistical differences were assessed using the Mann Whitney U test for two group comparison or the Kruskal Wallis test followed by pairwise comparison with an adjusted p value for more than two group comparisons. Differences at p < 0.05 was considered to be significant. The statistical analysis was performed using Prism 8 (GraphPad Software, CA, USA).
References
Canalis, E. et al. Notch signaling in osteocytes differentially regulates cancellous and cortical bone remodeling. J. Biol. Chem.288, 25614–25625 (2013).
Regan, J. & Long, F. Notch signaling and bone remodeling. Curr. Osteoporos. Rep.11, 126–129 (2013).
Kikuta, J., Yamaguchi, M., Shimizu, M., Yoshino, T. & Kasai, K. Notch signaling induces root resorption via RANKL and IL-6 from hPDL cells. J. Dent. Res.94, 140–147 (2015).
Manokawinchoke, J., Sumrejkanchanakij, P., Subbalekha, K., Pavasant, P. & Osathanon, T. Jagged1 inhibits osteoprotegerin expression by human periodontal ligament cells. J. Periodontal. Res.51, 789–799 (2016).
Nakao, A. et al. PTHrP induces Notch signaling in periodontal ligament cells. J. Dent. Res.88, 551–556 (2009).
Tanabe, H. et al. Periostin associates with Notch1 precursor to maintain Notch1 expression under a stress condition in mouse cells. PLoS ONE5, e12234 (2010).
Osathanon, T., Nowwarote, N., Manokawinchoke, J. & Pavasant, P. bFGF and JAGGED1 regulate alkaline phosphatase expression and mineralization in dental tissue-derived mesenchymal stem cells. J. Cell Biochem.114, 2551–2561 (2013).
Osathanon, T. et al. Surface-bound orientated Jagged-1 enhances osteogenic differentiation of human periodontal ligament-derived mesenchymal stem cells. J. Biomed. Mater. Res. A101, 358–367 (2013).
Liu, L., Ling, J., Wei, X., Wu, L. & Xiao, Y. Stem cell regulatory gene expression in human adult dental pulp and periodontal ligament cells undergoing odontogenic/osteogenic differentiation. J. Endod.35, 1368–1376 (2009).
Li, Y., Li, S. Q., Gao, Y. M., Li, J. & Zhang, B. Crucial role of Notch signaling in osteogenic differentiation of periodontal ligament stem cells in osteoporotic rats. Cell Biol. Int.38, 729–736 (2014).
Hozumi, K. Distinctive properties of the interactions between Notch and Notch ligands. Dev. Growth Differ.62, 49–58 (2020).
Mitsiadis, T. A., Lardelli, M., Lendahl, U. & Thesleff, I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J. Cell Biol.130, 407–418 (1995).
Meester, J. A. N. et al. Overlapping but distinct roles for NOTCH receptors in human cardiovascular disease. Clin. Genet.95, 85–94 (2019).
Nowwarote, N. et al. Characterization of a bioactive Jagged1-coated polycaprolactone-based membrane for guided tissue regeneration. Arch. Oral Biol.88, 24–33 (2018).
Zhu, F., Sweetwyne, M. T. & Hankenson, K. D. PKCdelta is required for Jagged-1 induction of human mesenchymal stem cell osteogenic differentiation. Stem Cells31, 1181–1192 (2013).
Osathanon, T. et al. Notch signaling is involved in neurogenic commitment of human periodontal ligament-derived mesenchymal stem cells. Stem Cells Dev.22, 1220–1231 (2013).
Hansamuit, K., Osathanon, T. & Suwanwela, J. Effect of Jagged1 on the expression of genes in regulation of osteoblast differentiation and bone mineralization ontology in human dental pulp and periodontal ligament cells. J. Oral Biol. Craniofac. Res.10, 233–237 (2020).
Osathanon, T. et al. Jagged1 promotes mineralization in human bone-derived cells. Arch. Oral Biol.99, 134–140 (2019).
Manokawinchoke, J. et al. Indirect immobilized Jagged1 suppresses cell cycle progression and induces odonto/osteogenic differentiation in human dental pulp cells. Sci. Rep.7, 10124 (2017).
Dishowitz, M. I. et al. Jagged1 immobilization to an osteoconductive polymer activates the Notch signaling pathway and induces osteogenesis. J. Biomed. Mater. Res. A102, 1558–1567 (2014).
Osathanon, T., Manokawinchoke, J., Egusa, H. & Pavasant, P. Notch signaling partly regulates the osteogenic differentiation of retinoic acid-treated murine induced pluripotent stem cells. J. Oral. Sci.59, 405–413 (2017).
Shimizu, T. et al. Notch signaling induces osteogenic differentiation and mineralization of vascular smooth muscle cells: role of Msx2 gene induction via Notch-RBP-Jk signaling. Arterioscler. Thromb. Vasc. Biol.29, 1104–1111 (2009).
Anant, S., Roy, S. & VijayRaghavan, K. Twist and Notch negatively regulate adult muscle differentiation in Drosophila. Development125, 1361–1369 (1998).
Zhang, X. W. et al. Twist-related protein 1 negatively regulated osteoblastic transdifferentiation of human aortic valve interstitial cells by directly inhibiting runt-related transcription factor 2. J. Thorac. Cardiovasc. Surg.148, 1700–1708 (2014).
Komaki, M. et al. Twist negatively regulates osteoblastic differentiation in human periodontal ligament cells. J. Cell. Biochem.100, 303–314 (2007).
Manokawinchoke, J., Sumrejkanchanakij, P., Pavasant, P. & Osathanon, T. Notch signaling participates in TGF-beta-induced SOST expression under intermittent compressive stress. J. Cell Physiol.232, 2221–2230 (2017).
Baeten, J. T. & Lilly, B. Differential regulation of NOTCH2 and NOTCH3 contribute to their unique functions in vascular smooth muscle cells. J. Biol. Chem.290, 16226–16237 (2015).
Baumgart, A. et al. Opposing role of Notch1 and Notch2 in a Kras(G12D)-driven murine non-small cell lung cancer model. Oncogene34, 578–588 (2015).
Yorgan, T. et al. Osteoblast-specific Notch2 inactivation causes increased trabecular bone mass at specific sites of the appendicular skeleton. Bone87, 136–146 (2016).
Ongaro, A. et al. Characterization of notch signaling during osteogenic differentiation in human osteosarcoma cell line MG63. J. Cell Physiol.231, 2652–2663 (2016).
Chakravorty, N. et al. Pro-osteogenic topographical cues promote early activation of osteoprogenitor differentiation via enhanced TGFbeta, Wnt, and Notch signaling. Clin. Oral Implants Res.25, 475–486 (2014).
Hoac, B., Nelea, V., Jiang, W., Kaartinen, M. T. & McKee, M. D. Mineralization-inhibiting effects of transglutaminase-crosslinked polymeric osteopontin. Bone101, 37–48 (2017).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods25, 402–408 (2001).
Acknowledgements
This study was supported by Thailand Research Fund (RTA6180001 to P.P.) and the Faculty of Dentistry Research Funding, Chulalongkorn University (DRF 59011 to P.S.). T.O. is supported by the Thailand Research Fund (RSA6180019). The authors thank Dr. Kevin Tompkins for language editing and Immunology Research Center, Faculty of Dentistry, Chulalongkorn University for flow cytometry analysis.
Author information
Authors and Affiliations
Contributions
J.M. performed the cell culture experiments. L.B. contributed to the scanning electron microscope analysis. P.P., P.S., and H.E. contributed to data interpretation. T.O. contributed to conceptual design, data interpretation, and manuscript drafting. All authors critically revised the manuscript and gave final approval for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manokawinchoke, J., Sumrejkanchanakij, P., Boonprakong, L. et al. NOTCH2 participates in Jagged1-induced osteogenic differentiation in human periodontal ligament cells. Sci Rep 10, 13329 (2020). https://doi.org/10.1038/s41598-020-70277-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70277-7
This article is cited by
-
Fibulin2: a negative regulator of BMSC osteogenic differentiation in infected bone fracture healing
Experimental & Molecular Medicine (2023)
-
Dissecting specific Wnt components governing osteogenic differentiation potential by human periodontal ligament stem cells through interleukin-6
Scientific Reports (2023)
-
Non-canonical Wnt signaling participates in Jagged1-induced osteo/odontogenic differentiation in human dental pulp stem cells
Scientific Reports (2022)
-
Phenytoin Regulates Migration and Osteogenic Differentiation by MAPK Pathway in Human Periodontal Ligament Cells
Cellular and Molecular Bioengineering (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.