Abstract
Huntington’s disease (HD) is a neurodegenerative disorder caused by expansion of a CAG trinucleotide repeat in the Huntingtin (HTT) gene, encoding a homopolymeric polyglutamine (polyQ) tract. Although mutant HTT (mHTT) protein is known to aggregate, the links between aggregation and neurotoxicity remain unclear. Here we show that both translation and aggregation of wild-type HTT and mHTT are regulated by a stress-responsive upstream open reading frame and that polyQ expansions cause abortive translation termination and release of truncated, aggregation-prone mHTT fragments. Notably, we find that mHTT depletes translation elongation factor eIF5A in brains of symptomatic HD mice and cultured HD cells, leading to pervasive ribosome pausing and collisions. Loss of eIF5A disrupts homeostatic controls and impairs recovery from acute stress. Importantly, drugs that inhibit translation initiation reduce premature termination and mitigate this escalating cascade of ribotoxic stress and dysfunction in HD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Previously published ribosome profiling analyses were from GSE87328 (Fig. 1c), GSE72064 (Fig. 1d,e), SRA160745, GSE74279 and GSE94460 (Extended Data Fig. 1a), GSE66715, GSE81283 and GSE36892 (Extended Data Fig. 1b) and GSE53743 and GSE119615 (Extended Data Fig. 1c). For analysis of previously published proteomic data, processed MS intensity tables were downloaded from the supplementary information section of the following published articles: Hosp et al.6, Supplementary Tables 1 and 3 (label-free quantification, LFQ); Sap et al.50, Supplementary Tables 1 and 3 (LFQ); Newcombe et al.43, Supplementary Table 2 (LFQ); and Sui et al.45, Supplementary Table 1 (raw intensities). No additional normalization of the data was performed. Statistical tests were done using two-sided Student’s t-test. Sequencing data were deposited in the Sequence Read Archive (SRA) database under BioProject number PRJNA730032. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE72 partner repository with the dataset identifier PXD026012. Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Bates, G. P. et al. Huntington disease. Nat. Rev. Dis. Prim. 1, 15005 (2015).
Finkbeiner, S. Huntington’s disease. Cold Spring Harb. Perspect. Biol. 3, a007476 (2011).
Paulson, H. L., Shakkottai, V. G., Clark, H. B. & Orr, H. T. Polyglutamine spinocerebellar ataxias—from genes to potential treatments. Nat. Rev. Neurosci. 18, 613–626 (2017).
Jimenez-Sanchez, M., Licitra, F., Underwood, B. R. & Rubinsztein, D. C. Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb. Perspect. Med. 7, a024240 (2017).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011).
Hosp, F. et al. Spatiotemporal proteomic profiling of Huntington’s disease inclusions reveals widespread loss of protein function. Cell Rep. 21, 2291–2303 (2017).
Kim, Y. E. et al. Soluble oligomers of polyQ-expanded Huntingtin target a multiplicity of key cellular factors. Mol. Cell 63, 951–964 (2016).
Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004).
Nalavade, R., Griesche, N., Ryan, D. P., Hildebrand, S. & Krauß, S. Mechanisms of RNA-induced toxicity in CAG repeat disorders. Cell Death Dis. 4, e752 (2013).
Lee, J.-M. et al. CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900.e14 (2019).
Lee, J. et al. An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res. 30, 5110–5119 (2002).
Kozak, M. The scanning model for translation: an update. J. Cell Biol. 108, 229–241 (1989).
Zhang, H., Wang, Y. & Lu, J. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem. Sci. 44, 782–794 (2019).
Fijałkowska, D. et al. eIF1 modulates the recognition of suboptimal translation initiation sites and steers gene expression via uORFs. Nucleic Acids Res. 45, 7997–8013 (2017).
Cho, J. et al. Multiple repressive mechanisms in the hippocampus during memory formation. Science 350, 82–87 (2015).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Rodriguez, C. M., Chun, S. Y., Mills, R. E. & Todd, P. K. Translation of upstream open reading frames in a model of neuronal differentiation. BMC Genomics 20, 391 (2019).
Young, S. K. & Wek, R. C. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 291, 16927–16935 (2016).
Reid, D. W. et al. Complementary roles of GADD34- and CReP-containing eukaryotic initiation factor 2α phosphatases during the unfolded protein response. Mol. Cell. Biol. 36, 1868–1880 (2016).
Trettel, F. Dominant phenotypes produced by the HD mutation in STHdhQ111 striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000).
Juszkiewicz, S. & Hegde, R. S. Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4 (2017).
Juszkiewicz, S. et al. Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. eLife 9, e60038 (2020).
Ferrin, M. A. & Subramaniam, A. R. Kinetic modeling predicts a stimulatory role for ribosome collisions at elongation stall sites in bacteria. eLife 6, e23629 (2017).
Creus-Muncunill, J. et al. Increased translation as a novel pathogenic mechanism in Huntington’s disease. Brain 142, 3158–3175 (2019).
Yip, M. C. J. & Shao, S. Detecting and rescuing stalled ribosomes. Trends Biochem. Sci. 46, 731–743 (2021).
Zhao, T. et al. Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding. Genome Biol. 22, 16 (2021).
Doerfel, L. K. et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339, 85–88 (2013).
Huter, P. et al. Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol. Cell 68, 515–527.e6 (2017).
Gingras, A. C., Svitkin, Y., Belsham, G. J., Pause, A. & Sonenberg, N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl Acad. Sci. USA 93, 5578–5583 (1996).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Arpat, A. B. et al. Transcriptome-wide sites of collided ribosomes reveal principles of translational pausing. Genome Res. 30, 985–999 (2020).
Sinha, N. K. et al. EDF1 coordinates cellular responses to ribosome collisions. eLife 9, e58828 (2020).
Pochopien, A. A. et al. Structure of Gcn1 bound to stalled and colliding 80S ribosomes. Proc. Natl Acad. Sci. USA 118, e2022756118 (2021).
Koyuncu, S. et al. The ubiquitin ligase UBR5 suppresses proteostasis collapse in pluripotent stem cells from Huntington’s disease patients. Nat. Commun. 9, 2886 (2018).
Ghosh, D. K., Roy, A. & Ranjan, A. The ATPase VCP/p97 functions as a disaggregase against toxic Huntingtin‐exon1 aggregates. FEBS Lett. 592, 2680–2692 (2018).
Zhou, M. et al. HUWE1 amplifies ubiquitin modifications to broadly stimulate clearance of proteins and aggregates. Preprint at bioRxiv https://doi.org/10.1101/2023.05.30.542866 (2023).
Huang, Z.-N. & Her, L.-S. The ubiquitin receptor ADRM1 modulates HAP40-induced proteasome activity. Mol. Neurobiol. 54, 7382–7400 (2017).
Wyttenbach, A. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum. Mol. Genet. 11, 1137–1151 (2002).
Pelechano, V. & Alepuz, P. eIF5A facilitates translation termination globally and promotes the elongation of many non-polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338 (2017).
Manjunath, H. et al. Suppression of ribosomal pausing by eIF5A is necessary to maintain the fidelity of start codon selection. Cell Rep. 29, 3134–3146.e6 (2019).
Faundes, V. et al. Impaired eIF5A function causes a Mendelian disorder that is partially rescued in model systems by spermidine. Nat. Commun. 12, 833 (2021).
Smeltzer, S. et al. Hypusination of Eif5a regulates cytoplasmic TDP-43 aggregation and accumulation in a stress-induced cellular model. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 165939 (2021).
Newcombe, E. A. et al. Tadpole-like conformations of Huntingtin exon 1 are characterized by conformational heterogeneity that persists regardless of polyglutamine length. J. Mol. Biol. 430, 1442–1458 (2018).
Gruber, A. et al. Molecular and structural architecture of polyQ aggregates in yeast. Proc. Natl Acad. Sci. USA 115, E3446–E3453 (2018).
Sui, X. et al. Widespread remodeling of proteome solubility in response to different protein homeostasis stresses. Proc. Natl Acad. Sci. USA 117, 2422–2431 (2020).
Schuller, A. P., Wu, C. C.-C., Dever, T. E., Buskirk, A. R. & Green, R. eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5 (2017).
Han, P. et al. Genome-wide survey of ribosome collision. Cell Rep. 31, 107610 (2020).
Higgins, R. et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell 59, 35–49 (2015).
Garshott, D. M., Sundaramoorthy, E., Leonard, M. & Bennett, E. J. Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. eLife 9, e54023 (2020).
Sap, K. A. et al. Global proteome and ubiquitinome changes in the soluble and insoluble fractions of Q175 Huntington mice brains. Mol. Cell. Proteom. 18, 1705–1720 (2019).
Eshraghi, M. et al. Mutant Huntingtin stalls ribosomes and represses protein synthesis in a cellular model of Huntington disease. Nat. Commun. 12, 1461 (2021).
Stein, K. C., Morales-Polanco, F., van der Lienden, J., Rainbolt, T. K. & Frydman, J. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 601, 637–642 (2022).
Zavodszky, E., Peak-Chew, S.-Y., Juszkiewicz, S., Narvaez, A. J. & Hegde, R. S. Identification of a quality-control factor that monitors failures during proteasome assembly. Science 373, 998–1004 (2021).
Shacham, T., Sharma, N. & Lederkremer, G. Z. Protein misfolding and ER stress in Huntington’s disease. Front. Mol. Biosci. 6, 20 (2019).
Duennwald, M. L. & Lindquist, S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 22, 3308–3319 (2008).
Wu, C. C.-C., Peterson, A., Zinshteyn, B., Regot, S. & Green, R. Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182, 404–416.e14 (2020).
Glock, C., Heumüller, M. & Schuman, E. M. mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 45, 169–177 (2017).
Sathasivam, K. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. USA 110, 2366–2370 (2013).
Goldberg, Y. P. et al. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat. Genet. 13, 442–449 (1996).
Bañez-Coronel, M. et al. RAN translation in Huntington disease. Neuron 88, 667–677 (2015).
Smith, A. M., Costello, M. S., Kettring, A. H., Wingo, R. J. & Moore, S. D. Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs. Proc. Natl Acad. Sci. USA 116, 21769–21779 (2019).
Simms, C. L., Yan, L. L., Qiu, J. K. & Zaher, H. S. Ribosome collisions result in +1 frameshifting in the absence of no-go decay. Cell Rep. 28, 1679–1689.e4 (2019).
Michel, A. M. et al. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 42, D859–D864 (2014).
Wang, H. et al. RPFdb v2.0: an updated database for genome-wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Res. 47, D230–D234 (2019).
Duim, W. C., Jiang, Y., Shen, K., Frydman, J. & Moerner, W. E. Super-resolution fluorescence of Huntingtin reveals growth of globular species into short fibers and coexistence of distinct aggregates. ACS Chem. Biol. 9, 2767–2778 (2014).
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M. & Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Kumari, R., Michel, A. M. & Baranov, P. V. PausePred and Rfeet: webtools for inferring ribosome pauses and visualizing footprint density from ribosome profiling data. RNA 24, 1297–1304 (2018).
Reinhart, P. H. et al. Identification of anti-inflammatory targets for Huntington’s disease using a brain slice-based screening assay. Neurobiol. Dis. 43, 248–256 (2011).
Xiao, Y. et al. Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat. Commun. 8, 375 (2017).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Acknowledgements
We thank I. Munoz-Sanjuan and D. Marchionini, as well as members of the Frydman lab, for their helpful comments. This work was supported by NIH grants GM05643321, NS092525 to J.F. and AI36178, AI40085 and AI091575 to R. Andino and Cure Huntington’s Disease Initiative (CHDI) grant A-13887 to J.F.
Author information
Authors and Affiliations
Contributions
R. Aviner and J.F. designed the study. R. Aviner and T.-T.L. carried out experiments, analysed data and performed statistical analyses. K.H.L. performed LC–MS/MS data acquisition. V.B.M. performed cloning. R. Aviner, R. Andino and J.F. wrote the manuscript, and all authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
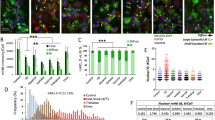
Extended Data Fig. 1 HTT translation initiation is dynamically regulated by its uORF.
(a) Ribo-seq traces of initiating ribosomes (A-sites, top) and elongating ribosomes (footprints, bottom) on human HTT uORF and start of main ORF. (b) Top, ribo-seq traces of elongating ribosomes on Htt uORF and start of main ORF in different organisms and tissues. Bottom, Htt protein level (low, moderate, high) in the indicated tissues, from ProteinAtlas. (c) Ribo-seq traces of elongating ribosomes on Htt and Atf4 mRNA in mouse embryonic fibroblasts before and after thapsigargin (Tg) treatment (left, n = 2) and SH-SY5Y neuroblasts before and after differentiation with retinoic acid (right, n = 2).
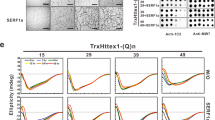
Extended Data Fig. 2 HTT uORF reduces truncation and aggregation of mHTT.
(a) 293 T cells were transfected with constructs expressing wild-type polyQ(25) or mutant polyQ(97) HTT-ex1 with a C-terminal fluorescent GFP. Shown are live cell images at the indicated times post-transfection. Images were obtained using 20x magnification. Representative images of 2 independent replicates. (b-c) Immunoblot analyses of 293 T (b) and mouse striatal cells (c) transfected as above and separated into soluble and insoluble fractions (n = 2). WB, western blot. (d) Flow cytometry analysis of transfected 293 T cells. (e) Effect of 4EGI-1 on global translation. 293 T cells were treated with increasing concentrations of 4EGI-1 for 24 h, followed by puromycin for 15 min (n = 2). Immunoblot analysis was performed on whole cell lysates using a puromycin-specific antibody. (f) 293 T cells were transfected with mHTT-ex1-GFP and increasing concentrations of 4EGI-1 were added 6 h post-transfection. Immunoblot analysis of the insoluble fraction was performed at 24 h post-transfection (n = 3). Means +/- s.d. P, p-value of a two-tailed Student t-test comparing 50 and 0 µM 4EGI-1. Unprocessed blots are available in source data.
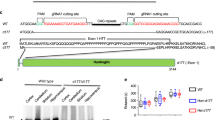
Extended Data Fig. 3 Translation of mHTT from poliovirus genome is associated with ribosome collisions.
(a) Poliovirus replicates more slowly when engineered to express mutant versus wild-type HTT-ex1. Wild-type polyQ(8) and mutant polyQ(73) human HTT-ex1 were cloned into poliovirus genome. Infectious viruses were generated by transfection of in vitro transcribed RNA. Plaque assays were performed to determine virus titers (n = 3). Means +/- s.d. (left) and representative images of viral plaques (right). (b-c) Huh7 cells (b) and SH-SY5Y neuroblasts (c) were infected with engineered viruses. At the indicated times, puromycin was added to tissue culture media to label nascent polypeptide chains and lysates were analyzed by immunoblotting. Shown is a representative of 2 independent repeats. Purple boxes and arrowheads indicate virus-Htt polyproteins prior to cleavage by the viral protease. WB, western blot. (d) Quantification of blots from Fig. 3c (n = 3). Means +/- s.d. (e) Huh7 cells were infected with HTT-ex1-expressing viruses for 3.5 h, and ribosomes were isolated by ultracentrifugation on a sucrose cushion, followed by immunoblot analysis using antibodies specific to viral epitopes upstream and downstream of HTT-ex1 insert (n = 2). Unprocessed blots are available in source data.
Extended Data Fig. 4 HD mouse brains show age-dependent changes in levels of soluble translation and RQC factors.
(a) eIF5A, but not other elongation factors, is inversely correlated with mHTT levels in mouse brains at 12 weeks of age. Pearson’s correlation coefficients for wild-type or mutant Htt and the indicated translation elongation factors. Reanalysis of published MS analysis of soluble brain proteomes from 12-week-old R6/2 and control mice (n = 16/12). (b-c) Levels of RQC factors increase, while those of elongation factors decrease, in the soluble brain proteome of presymptomatic R6/2 HD mice. Shown are levels of the indicated proteins in wild-type and R6/2 HD cells at 5, 8 and 12 weeks of age (b) and their Student’s t-test differences between wild-type and R6/2 at 8 weeks. n = 16, 12 and 12 for WT or n = 16, 12 and 16 for R6/2 at weeks 5, 8 and 12, respectively. Means +/-s.d. (c). Histogram shows the distribution of t-test differences of the entire soluble proteome at 8 weeks.
Extended Data Fig. 5 Disruption of proteostasis machines in R6/2 mice.
(a) Proteins encoded by pause-containing transcripts detected in striatal cells expressing polyQ(111) mHtt are enriched in the insoluble brain proteome of aged 12-week-old R6/2 mice (n = 12/16 for WT and R6/2, respectively). (b) Kurtosis is a measure of tailedness or outliers relative to a normal distribution, and can be used to estimate stoichiometry of large molecular complexes. (c-d) Stoichiometry of ribosomal proteins (c) and proteasomal subunits (d) is disrupted in R6/2 brains starting at 8 weeks of age. Shown are kurtosis scores for all core ribosomal proteins or proteasomal subunits as measured by MS. n = 16, 12 and 12 for WT or n = 16, 12 and 16 for R6/2 at weeks 5, 8 and 12, respectively. (e) Ribosomal proteins are enriched in the insoluble brain proteome of symptomatic 12-week-old but not asymptomatic 8-week-old R6/2 mice. n = 16, 12 and 12 for WT or n = 16, 12 and 16 for R6/2 at weeks 5, 8 and 12, respectively. Proteasomal subunits were not detected in the insoluble brain proteome of either wild-type or R6/2 mice at any age. For a, c-e, center lines show medians; box limits—upper and lower quartiles; and whiskers extend 1.5 times the interquartile range.
Extended Data Fig. 6 Global and Atf4-specific translation levels in mouse striatal cells.
(a) Global translation is lower in striatal cells expressing polyQ(111) mHtt. Translation rates were monitored by puromycin labeling and quantified by densitometry (left, n = 3, Means +/-s.d.) and polysome profiles on 10-50% sucrose gradients (right, n = 2, representative traces). WB, western blot. (b) Translation efficiency of Atf4 is lower in polyQ(111) cells at steady-state. Means of values from RNA- and ribo-seq analyses (n = 2 each). (c) Ribo-seq traces of elongating ribosomes on Atf4 mRNA (n = 2). Unprocessed blots are available in source data.
Supplementary information
Supplementary Table 1-6
MS data, ribosome stalling analyses and plasmid/oligonucleotide information.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Uncropped blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aviner, R., Lee, TT., Masto, V.B. et al. Polyglutamine-mediated ribotoxicity disrupts proteostasis and stress responses in Huntington’s disease. Nat Cell Biol (2024). https://doi.org/10.1038/s41556-024-01414-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41556-024-01414-x