Abstract
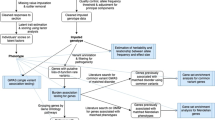
Mental health conditions are characterized by higher-order transdiagnostic factor structures, which may contribute to the high levels of comorbidity observed in psychopathology. However, the phenotypic and genetic structures of various psychopathology diagnoses may differ, raising questions about the validity and utility of these factors. Here we study the phenotypic and genetic factor structures of ten psychiatric conditions using UK Biobank and public genomic data. Although the factor structure of psychopathology was generally genetically and phenotypically consistent, conditions related to externalizing (for example, alcohol use disorder) and compulsivity (for example, eating disorders) exhibited cross-level disparities in their relationships with other conditions, possibly due to environmental influences. Domain-level factors, especially thought disorder and internalizing factors, were more informative than a general psychopathology factor in genome-wide association and polygenic index analyses. Collectively, our findings enhance the understanding of comorbidity and shared etiology, highlight the intricate interplay between genes and environment, and offer guidance for psychiatric research using polygenic indices.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$79.00 per year
only $6.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
No new data were gathered for this study; instead, data from pre-existing studies or cohorts were utilized. The essential dataset needed to understand, replicate, and expand on the findings, specifically the GWAS summary statistics for the psychopathology GWAS, are available at an Open Science Framework repository (https://osf.io/unkym/). The UK Biobank and the Mass General Brigham Biobank have access restrictions in place to safeguard participant privacy. The UK Biobank data are accessible to researchers with an ongoing application in the UK Biobank (https://www.ukbiobank.ac.uk/). This study was conducted on the basis of the UK Biobank application 46007. We used reference data from the 1000 Genomes phase 3 (version 5) (https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html) and HapMap 3 (revision 2) (https://mathgen.stats.ox.ac.uk/impute/data_download_hapmap3_r2.html). Publicly available GWAS summary statistics included in the meta-analyses include alcohol use disorder (https://pubmed.ncbi.nlm.nih.gov/29058377/), anorexia (https://doi.org/10.1176/appi.ajp.2017.16121402), generalized anxiety (https://jamanetwork.com/journals/jamapsychiatry/fullarticle/2733149), bipolar disorder (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6956732/), depression (https://www.nature.com/articles/s41588-018-0090-3), suicide attempt (https://pubmed.ncbi.nlm.nih.gov/30116032/), obsessive–compulsive disorder (https://pubmed.ncbi.nlm.nih.gov/28761083/), panic disorder (https://www.nature.com/articles/s41380-019-0590-2), post-traumatic stress disorder (https://pubmed.ncbi.nlm.nih.gov/28439101/), and schizophrenia (https://pubmed.ncbi.nlm.nih.gov/28439101/). Cortical maps were obtained from the neuromaps toolbox (https://www.nature.com/articles/s41592-022-01625-w). Gene expression datasets from GTEx8 and Brainspan are available through FUMA (https://fuma.ctglab.nl/).
Code availability
The code for the present study is available on OSF (https://osf.io/unkym/). This wraps fastGWA-GLMM from the GCTA tool (version 1.94.1) (https://yanglab.westlake.edu.cn/software/gcta/#fastGWA-GLMM), Genomic SEM (version 0.0.5) (https://github.com/GenomicSEM/GenomicSEM), METAL (version 2020-05-05) (https://genome.sph.umich.edu/wiki/METAL_Documentation), PRS-CS (version October 20, 2019) (https://github.com/getian107/PRScs), sBayesR (version 2.05) (https://cnsgenomics.com/software/gctb/#Overview), plink package (version 1.5.1) (https://www.cog-genomics.org/plink/1.9/rserve), and R (version 4.1.0) (https://cran.rstudio.com/). Bioannotation analyses were conducted using FUMA (version 1.3.5e) (https://fuma.ctglab.nl/).
Change history
09 July 2024
A Correction to this paper has been published: https://doi.org/10.1038/s44220-024-00293-3
References
Kessler, R. C., Chiu, W. T., Demler, O. & Walters, E. E. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry 62, 617–627 (2005).
Klein Hofmeijer-Sevink, M. et al. Clinical relevance of comorbidity in anxiety disorders: a report from the Netherlands Study of Depression and Anxiety (NESDA). J. Affect. Disord. 137, 106–112 (2012).
Investing in Mental Health (WHO, 2003); https://apps.who.int/iris/bitstream/handle/10665/42823/9241562579.pdf
Insel, T. et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry 167, 748–751 (2010).
Kotov, R. et al. The Hierarchical Taxonomy of Psychopathology (HiTOP): a dimensional alternative to traditional nosologies. J. Abnorm. Psychol. 126, 454–477 (2017).
Beam, E., Potts, C., Poldrack, R. A. & Etkin, A. A data-driven framework for mapping domains of human neurobiology. Nat. Neurosci. 24, 1733–1744 (2021).
Dalgleish, T., Black, M., Johnston, D. & Bevan, A. Transdiagnostic approaches to mental health problems: current status and future directions. J. Consult. Clin. Psychol. 88, 179–195 (2020).
Lee, P. H. et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019).
Grotzinger, A. D. et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat. Genet. 54, 548–559 (2022).
Mallard, T. T., Grotzinger, A. D. & Smoller, J. W. Examining the shared etiology of psychopathology with genome-wide association studies. Physiol. Rev. 103, 1645–1665 (2023).
Smoller, J. W. et al. Psychiatric genetics and the structure of psychopathology. Mol. Psychiatry 24, 409–420 (2019).
Polderman, T. J. C. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Jones, H. J. et al. Investigating the genetic architecture of general and specific psychopathology in adolescence. Transl. Psychiatry 8, 145 (2018).
The Brainstorm Consortium et al. Analysis of shared heritability in common disorders of the brain. Science 360, eaap8757 (2018).
Caspi, A. et al. The p factor: one general psychopathology factor in the structure of psychiatric disorders? Clin. Psychol. Sci. 2, 119–137 (2014).
Lahey, B. B. et al. Measuring the hierarchical general factor model of psychopathology in young adults. Int. J. Methods Psychiatr. Res. 27, e1593 (2018).
Markon, K. E. Modeling psychopathology structure: a symptom-level analysis of Axis I and II disorders. Psychol. Med. 40, 273–288 (2010).
Markon, K. E. Bifactor and hierarchical models: specification, inference, and interpretation. Annu. Rev. Clin. Psychol. 15, 51–69 (2019).
Sprooten, E., Franke, B. & Greven, C. U. The P-factor and its genomic and neural equivalents: an integrated perspective. Mol. Psychiatry 27, 38–48 (2021).
Carey, C. E. et al. Principled distillation of multidimensional UK Biobank data reveals insights into the correlated human phenome. Preprint at medRxiv https://doi.org/10.1101/2022.09.02.22279546 (2022)
Caspi, A. et al. Longitudinal assessment of mental health disorders and comorbidities across 4 decades among participants in the Dunedin Birth Cohort Study. JAMA Netw. Open 3, e203221 (2020).
Caspi, A. & Moffitt, T. E. All for one and one for all: mental disorders in one dimension. Am. J. Psychiatry 175, 831–844 (2018).
Lahey, B. B., Krueger, R. F., Rathouz, P. J., Waldman, I. D. & Zald, D. H. Validity and utility of the general factor of psychopathology. World Psychiatry 16, 142–144 (2017).
Lahey, B. B., Moore, T. M., Kaczkurkin, A. N. & Zald, D. H. Hierarchical models of psychopathology: empirical support, implications, and remaining issues. World Psychiatry 20, 57–63 (2021).
Watts, A. L., Greene, A. L., Bonifay, W. & Fried, E. I. A critical evaluation of the p-factor literature. Nat. Rev. Psychol. 3, 108–122 (2024).
van Bork, R. et al. Latent variable models and networks: statistical equivalence and testability. Multivar. Behav. Res. 56, 175–198 (2019).
Waldman, I. D. et al. Recommendations for adjudicating among alternative structural models of psychopathology. Clin. Psychol. Sci. 11, 616–640 (2023).
Watts, A. L., Lane, S. P., Bonifay, W., Steinley, D. & Meyer, F. A. C. Building theories on top of, and not independent of, statistical models: the case of the p-factor. Psychol. Inq. 31, 310–320 (2020).
Clapp Sullivan, M. L. et al. Beyond the factor indeterminacy problem using genome-wide association data. Nat. Hum. Behav. 8, 205–218 (2024).
Vandekerckhove, J., Matzke, D. & Wagenmakers, E.-J. Model comparison and the principle of parsimony. In The Oxford Handbook of Computational and Mathematical Psychology (eds. Busemeyer, J. R., Wang, Z., Townsend, J. T. & Eidels, A.) 300–319 (Oxford Univ. Press, 2015).
Sodini, S. M., Kemper, K. E., Wray, N. R. & Trzaskowski, M. Comparison of genotypic and phenotypic correlations: Cheverud’s conjecture in humans. Genetics 209, 941–948 (2018).
Willis, J. H., Coyne, J. A. & Kirkpatrick, M. Can one predict the evolution of quantitative characters without genetics? Evolution 45, 441–444 (1991).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Karlsson Linnér, R. et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat. Neurosci. 24, 1367–1376 (2021).
Williams, C. M. et al. Guidelines for evaluating the comparability of down-sampled GWAS summary statistics. Behav. Genet. 53, 404–415 (2023).
Mallard, T. T. et al. Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genomics 2, 100140 (2022).
Sydnor, V. J. et al. Neurodevelopment of the association cortices: patterns, mechanisms, and implications for psychopathology. Neuron 109, 2820–2846 (2021).
Markello, R. D. et al. neuromaps: structural and functional interpretation of brain maps. Nat. Methods 19, 1472–1479 (2022).
Karlson, E. W., Boutin, N. T., Hoffnagle, A. G. & Allen, N. L. Building the Partners HealthCare Biobank at Partners Personalized Medicine: informed consent, return of research results, recruitment lessons and operational considerations. J. Pers. Med. 6, 2 (2016).
Cheverud, J. M. A comparison of genetic and phenotypic correlations. Evolution 42, 958–968 (1988).
Turkheimer, E., Pettersson, E. & Horn, E. E. A phenotypic null hypothesis for the genetics of personality. Annu. Rev. Psychol. 65, 515–540 (2014).
Weiner, D. J. et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614, 492–499 (2023).
Xie, C. et al. A shared neural basis underlying psychiatric comorbidity. Nat. Med. 29, 1232–1242 (2023).
Goodkind, M. et al. Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry 72, 305–315 (2015).
Taylor, J. J. et al. A transdiagnostic network for psychiatric illness derived from atrophy and lesions. Nat. Hum. Behav. 7, 420–429 (2023).
Grotzinger, A. D. et al. Multivariate genomic architecture of cortical thickness and surface area at multiple levels of analysis. Nat. Commun. 14, 946 (2023).
Cai, N. et al. Minimal phenotyping yields genome-wide association signals of low specificity for major depression. Nat. Genet. 52, 437–447 (2020).
Bulik-Sullivan, B. K. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Ruan, Y. et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet. 54, 573–580 (2022).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Abdellaoui, A., Smit, D. J. A., Brink, W., van den, Denys, D. & Verweij, K. J. H. Genomic relationships across psychiatric disorders including substance use disorders. Drug Alcohol Depend. 220, 108535 (2021).
Thorp, J. G. et al. Symptom-level modelling unravels the shared genetic architecture of anxiety and depression. Nat. Hum. Behav. 5, 1432–1442 (2021).
Abdellaoui, A. The evolutionary dance between culture, genes, and everything in between. Behav. Brain Sci. 45, e153 (2022).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Alfaro-Almagro, F. et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage 166, 400–424 (2018).
Miller, K. L. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 19, 1523–1536 (2016).
Williams, C. M. et al. High intelligence is not associated with a greater propensity for mental health disorders. Eur. Psychiatry 66, e3 (2023).
R. Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
Revelle, W. Psych: Procedures for Psychological, Psychometric, and Personality Research (Northwestern Univ., 2021).
Finch, W. H. Using fit statistic differences to determine the optimal number of factors to retain in an exploratory factor analysis. Educ. Psychol. Meas. 80, 217–241 (2020).
Fabrigar, L. R., Wegener, D., MacCallum, R. C. & Strahan, E. J. Evaluating the use of exploratory factor analysis in psychological research. Psychol. Methods 4, 272–299 (1999).
Rosseel, Y. lavaan: an R package for structural equation modeling. J. Stat. Softw. 48, 1–36 (2012).
Kolenikov, S. & Bollen, K. A. Testing negative error variances: is a Heywood case a symptom of misspecification? Sociol. Methods Res. 41, 124–167 (2012).
Kline, R. B. Principles and Practice of Structural Equation Modeling 4th edn (Guilford Publications, 2015).
Marsh, H. W., Hau, K.-T. & Wen, Z. In search of golden rules: comment on hypothesis-testing approaches to setting cutoff values for fit indexes and dangers in overgeneralizing Hu and Bentler’s (1999) findings. Struct. Equ. Model. 11, 320–341 (2004).
Jiang, L., Zheng, Z., Fang, H. & Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 53, 1616–1621 (2021).
Yang, J., Lee, S. H., Wray, N. R., Goddard, M. E. & Visscher, P. M. GCTA-GREML accounts for linkage disequilibrium when estimating genetic variance from genome-wide SNPs. Proc. Natl Acad. Sci. USA 113, E4579–E4580 (2016).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
de la Fuente, J., Davies, G., Grotzinger, A. D., Tucker-Drob, E. M. & Deary, I. J. A general dimension of genetic sharing across diverse cognitive traits inferred from molecular data. Nat. Hum. Behav. 5, 49–58 (2021).
McArdle, J. J. & Goldsmith, H. H. Alternative common factor models for multivariate biometric analyses. Behav. Genet. 20, 569–608 (1990).
Lloyd-Jones, L. R. et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 10, 5086 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Hu, L. & Bentler, P. M. Cutoff criteria for fit indexes in covariance structure analysis: conventional criteria versus new alternatives. Struct. Equ. Model. 6, 1–55 (1999).
Boutin, N. T. et al. The evolution of a large biobank at Mass General Brigham. J. Pers. Med. 12, 1323 (2022).
Wei, W.-Q. et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE 12, e0175508 (2017).
Zheutlin, A. B. et al. Penetrance and pleiotropy of polygenic risk scores for schizophrenia in 106,160 patients across four health care systems. Am. J. Psychiatry 176, 846–855 (2019).
Carroll, R. J., Bastarache, L. & Denny, J. C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics 30, 2375–2376 (2014).
Alexander-Bloch, A. F. et al. On testing for spatial correspondence between maps of human brain structure and function. NeuroImage 178, 540–551 (2018).
Seidlitz, J. et al. Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron 97, 231–247.e7 (2018).
Váša, F. et al. Adolescent tuning of association cortex in human structural brain networks. Cereb. Cortex 28, 281–294 (2018).
Walters, R. K. et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669 (2018).
Meier, S. M. et al. Genetic variants associated with anxiety and stress-related disorders: a genome-wide association study and mouse-model study. JAMA Psychiatry 76, 924–932 (2019).
Forstner, A. J. et al. Genome-wide association study of panic disorder reveals genetic overlap with neuroticism and depression. Mol. Psychiatry 26, 4179–4190 (2021).
Stahl, E. A. et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 51, 793–803 (2019).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Duncan, L. et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858 (2017).
Arnold, P. D. et al. Revealing the complex genetic architecture of obsessive–compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188 (2018).
Duncan, L. E. et al. Largest GWAS of PTSD (N = 20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry 23, 666–673 (2018).
Pardiñas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Erlangsen, A. et al. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Mol. Psychiatry 25, 2410–2421 (2020).
Acknowledgments
We thank the UK Biobank staff, researchers, and volunteers. Funding: ANR-17-EURE-0017 (F.R.), ANR-10-IDEX-0001-02 PSL (F.R.), NIH T32HG010464 (T.T.M.), and NIH K08MH135343 (T.T.M.).
Author information
Authors and Affiliations
Contributions
C.M.W., H.P., F.R., and T.T.M. designed the study. T.G. conducted the polygenic score analyses with PRS-CS. Y.H.L. conducted the phenome-wide association analyses. J.S. conducted the correlational analyses of cortical maps. C.M.W. conducted all other analyses. T.W. and T.T.M. provided statistical guidance. C.M.W., J.W.S., F.R., T.G., and T.T.M. guided interpretation of key findings. All named authors reviewed, edited, and approved the submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Mental Health thanks Wei Cheng, Jeggan Tiego, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–34 and Sections 1–14.
Supplementary Data 1
Supplementary Tables 1–17.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Williams, C.M., Peyre, H., Wolfram, T. et al. Characterizing the phenotypic and genetic structure of psychopathology in UK Biobank. Nat. Mental Health 2, 960–974 (2024). https://doi.org/10.1038/s44220-024-00272-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44220-024-00272-8