Abstract
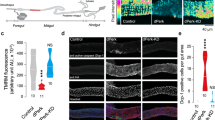
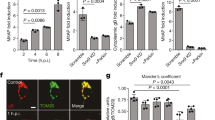
The innate immune response mounts a defense against foreign invaders and declines with age. An inappropriate induction of this response can cause diseases. Previous studies showed that mitochondria can be repurposed to promote inflammatory signaling. Damaged mitochondria can also trigger inflammation and promote diseases. Mutations in pink1, a gene required for mitochondrial health, cause Parkinson’s disease, and Drosophila melanogaster pink1 mutants accumulate damaged mitochondria. Here, we show that defective mitochondria in pink1 mutants activate Relish targets and demonstrate that inflammatory signaling causes age-dependent intestinal dysfunction in pink1-mutant flies. These effects result in the death of intestinal cells, metabolic reprogramming and neurotoxicity. We found that Relish signaling is activated downstream of a pathway stimulated by cytosolic DNA. Suppression of Relish in the intestinal midgut of pink1-mutant flies restores mitochondrial function and is neuroprotective. We thus conclude that gut–brain communication modulates neurotoxicity in a fly model of Parkinson’s disease through a mechanism involving mitochondrial dysfunction.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Source date files, including raw numerical data, descriptive statistics, normality tests and statistical analysis used in the manuscript, are available in our GitHub repository at github.com/M1gus/Gut-brain. Microarray data, with detailed descriptions of the experimental protocols, and raw data were deposited in ArrayExpress under accession no. E-MTAB-6210. Proteomics data was deposited at the ProteomeXchange Consortium with dataset identifier PXD030979. All other data are available upon reasonable request.
References
Collins, L. V., Hajizadeh, S., Holme, E., Jonsson, I. M. & Tarkowski, A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J. Leukoc. Biol. 75, 995–1000 (2004).
Celardo, I., Martins, L. M. & Gandhi, S. Unravelling mitochondrial pathways to Parkinson’s disease. Br. J. Pharmacol. 171, 1943–1957 (2014).
Oka, T. et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 485, 251–255 (2012).
Kounatidis, I. et al. NF-kappaB Immunity in the brain determines fly lifespan in healthy aging and age-related neurodegeneration. Cell Rep. 19, 836–848 (2017).
Myllymaki, H., Valanne, S. & Ramet, M. The Drosophila Imd signaling pathway. J. Immunol. 192, 3455–3462 (2014).
Molaei, M., Vandehoef, C. & Karpac, J. NF-kappaB shapes metabolic adaptation by attenuating Foxo-mediated lipolysis in Drosophila. Dev. Cell 49, 802–810 (2019).
Chinchore, Y., Gerber, G. F. & Dolph, P. J. Alternative pathway of cell death in Drosophila mediated by NF-kappaB transcription factor Relish. Proc. Natl Acad. Sci. USA 109, E605–E612 (2012).
Kuraishi, T., Hori, A. & Kurata, S. Host–microbe interactions in the gut of Drosophila melanogaster. Front. Physiol. 4, 375 (2013).
Sheldon, B. C. & Verhulst, S. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 11, 317–321 (1996).
Valtonen, T. M., Kleino, A., Ramet, M. & Rantala, M. J. Starvation reveals maintenance cost of humoral immunity. Evol. Biol. 37, 49–57 (2010).
Liu, X. et al. Drosophila EYA regulates the immune response against DNA through an evolutionarily conserved threonine phosphatase motif. PLoS ONE 7, e42725 (2012).
Tufi, R. et al. Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson’s disease. Nat. Cell Biol. 16, 157–166 (2014).
Celardo, I. et al. Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell Death Dis. 7, e2271 (2016).
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007).
Janky, R. et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014).
Julienne, H., Buhl, E., Leslie, D. S. & Hodge, J. J. L. Drosophila PINK1 and parkin loss-of-function mutants display a range of non-motor Parkinson’s disease phenotypes. Neurobiol. Dis. 104, 15–23 (2017).
Valadas, J. S. et al. ER lipid defects in neuropeptidergic neurons impair sleep patterns in Parkinson’s disease. Neuron 98, 1155–1169 (2018).
Park, J. et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 (2006).
Hedengren, M. et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4, 827–837 (1999).
Doktor, B., Damulewicz, M., Krzeptowski, W., Bednarczyk, B. & Pyza, E. M. Effects of PINK1 mutation on synapses and behavior in the brain of Drosophila melanogaster. Acta Neurobiol. Exp. (Wars.) 78, 231–241 (2018).
Li, Y. X. & Dijkers, P. F. Specific calcineurin isoforms are involved in Drosophila Toll immune signaling. J. Immunol. 194, 168–176 (2015).
Tain, L. S. et al. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 12, 1129–1135 (2009).
Han, Z. S. & Ip, Y. T. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 274, 21355–21361 (1999).
Franz, A., Wood, W. & Martin, P. Fat body cells are motile and actively migrate to wounds to drive repair and prevent infection. Dev. Cell 44, 460–470 (2018).
Karpac, J., Younger, A. & Jasper, H. Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev. Cell 20, 841–854 (2011).
Maitra, U., Scaglione, M. N., Chtarbanova, S. & O’Donnell, J. M. Innate immune responses to paraquat exposure in a Drosophila model of Parkinson’s disease. Sci. Rep. 9, 12714 (2019).
Rera, M., Clark, R. I. & Walker, D. W. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl Acad. Sci. USA 109, 21528–21533 (2012).
Suen, D. F., Norris, K. L. & Youle, R. J. Mitochondrial dynamics and apoptosis. Genes Dev. 22, 1577–1590 (2008).
Naszai, M., Carroll, L. R. & Cordero, J. B. Intestinal stem cell proliferation and epithelial homeostasis in the adult Drosophila midgut. Insect Biochem. Mol. Biol. 67, 9–14 (2015).
Micchelli, C. A. & Perrimon, N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479 (2006).
Harsh, S., Heryanto, C. & Eleftherianos, I. Intestinal lipid droplets as novel mediators of host–pathogen interaction in Drosophila. Biol. Open https://doi.org/10.1242/bio.039040 (2019).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Shokri, L. et al. A comprehensive Drosophila melanogaster transcription factor interactome. Cell Rep. 27, 955–970 (2019).
Sarov-Blat, L., So, W. V., Liu, L. & Rosbash, M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 101, 647–656 (2000).
Ahmad, M., He, L. & Perrimon, N. Regulation of insulin and adipokinetic hormone/glucagon production in flies. Wiley Interdiscip. Rev. Dev. Biol. https://doi.org/10.1002/wdev.360 (2019).
Post, S. et al. Drosophila insulin-like peptides DILP2 and DILP5 differentially stimulate cell signaling and glycogen phosphorylase to regulate longevity. Front. Endocrinol. (Lausanne) 9, 245 (2018).
Park, S. et al. A genetic strategy to measure circulating Drosophila insulin reveals genes regulating insulin production and secretion. PLoS Genet. 10, e1004555 (2014).
Heller, S. et al. Intestinal inflammation requires FOXO3 and prostaglandin E2-dependent lipogenesis and elevated lipid droplets. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G844–G854 (2016).
Kim, S. et al. Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627–641 (2019).
Miguel-Aliaga, I., Jasper, H. & Lemaitre, B. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics 210, 357–396 (2018).
Yin, S., Qin, Q. H. & Zhou, B. Functional studies of Drosophila zinc transporters reveal the mechanism for zinc excretion in Malpighian tubules. BMC Biol. 15, 12 (2017).
Nehme, N. T. et al. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3, 1694–1709 (2007).
Quinn, L. et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22, 3568–3579 (2003).
Allen, J. F. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631 (1993).
Sliter, D. A. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018).
Meyer, S. N. et al. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 346, 1258236 (2014).
Liu, Y. et al. Inflammation-induced, STING-dependent autophagy restricts Zika virus infection in the Drosophila brain. Cell Host Microbe 24, 57–68 (2018).
Lee, J. J., Andreazza, S. & Whitworth, A. J. The STING pathway does not contribute to behavioural or mitochondrial phenotypes in Drosophila Pink1/parkin or mtDNA mutator models. Sci. Rep. 10, 2693 (2020).
Petersen, A. J., Katzenberger, R. J. & Wassarman, D. A. The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics 194, 133–142 (2013).
Lee, H. S., Lobbestael, E., Vermeire, S., Sabino, J. & Cleynen, I. Inflammatory bowel disease and Parkinson’s disease: common pathophysiological links. Gut 70, 408–417 (2021).
Bravo-San Pedro, J. M. & Senovilla, L. Immunostimulatory activity of lifespan-extending agents. Aging (Albany NY) 5, 793–801 (2013).
Puig, O., Marr, M. T., Ruhf, M. L. & Tjian, R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006–2020 (2003).
Rynes, J. et al. Activating transcription factor 3 regulates immune and metabolic homeostasis. Mol. Cell. Biol. 32, 3949–3962 (2012).
Song, W. et al. Activin signaling mediates muscle-to-adipose communication in a mitochondria dysfunction-associated obesity model. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1708037114 (2017).
Van Den Berge, N. et al. Evidence for bidirectional and trans-synaptic parasympathetic and sympathetic propagation of alpha-synuclein in rats. Acta Neuropathol. 138, 535–550 (2019).
Scopelliti, A. et al. A neuronal relay mediates a nutrient ersponsive gut/fat body axis regulating energy homeostasis in adult Drosophila. Cell Metab. 29, 269–284 (2019).
Melnattur, K., Zhang, B. & Shaw, P. J. Disrupting flight increases sleep and identifies a novel sleep-promoting pathway in Drosophila. Sci. Adv. 6, eaaz2166 (2020).
Williams, J. A., Sathyanarayanan, S., Hendricks, J. C. & Sehgal, A. Interaction between sleep and the immune response in Drosophila: a role for the NFkappaB relish. Sleep 30, 389–400 (2007).
Badinloo, M. et al. Overexpression of antimicrobial peptides contributes to aging through cytotoxic effects in Drosophila tissues. Arch. Insect Biochem. Physiol. 98, e21464 (2018).
Clark, R. I. et al. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep. 12, 1656–1667 (2015).
Nassel, D. R., Kubrak, O. I., Liu, Y. T., Luo, J. N. & Lushchak, O. V. Factors that regulate insulin producing cells and their output in Drosophila. Front. Physiol. 4, 252 (2013).
Gatto, L. & Lilley, K. S. MSnbase—an R/bioconductor package for isobaric tagged mass spectrometry data visualization, processing and quantitation. Bioinformatics 28, 288–289 (2012).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995).
Donelson, N. C. et al. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS ONE 7, e37250 (2012).
Rera, M. et al. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 14, 623–634 (2011).
Martins, R. R., McCracken, A. W., Simons, M. J. P., Henriques, C. M. & Rera, M. How to catch a Smurf? – Ageing and beyond… in vivo assessment of intestinal permeability in multiple model organisms. Bio-Protoc. https://doi.org/10.21769/BioProtoc.2722 (2018).
Whitworth, A. J. et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl Acad. Sci. USA 102, 8024–8029 (2005).
Tennessen, J. M., Barry, W. E., Cox, J. & Thummel, C. S. Methods for studying metabolism in Drosophila. Methods 68, 105–115 (2014).
Moraru, A. et al. Elevated levels of the reactive metabolite methylglyoxal recapitulate progression of type 2 diabetes. Cell Metab. 27, 926–934 (2018).
Zhao, X. & Karpac, J. Muscle directs diurnal energy homeostasis through a myokine-dependent hormone module in Drosophila. Curr. Biol. 27, 1941–1955 (2017).
Wexler, E. J. Photoshop CS3 Extended for BioMedical Research CD-ROM – February 8, 2008 (Lynda.com, Inc., 2008).
Acknowledgements
Proteomics analysis was performed at the Cambridge Centre for Proteomics by R. Feret, M. Deery and Y. Umrania. We thank the Vienna Drosophila RNAi Center, Bloomington Drosophila Stock Center, J. Cordero, A. Telemans, A. Franz and E. Rosato for provision of fly stocks; the Fly Facility, Department of Genetics, University of Cambridge, for maintaining the stocks; and T. Ashby and M. Patel for preparing the fly food. This work was funded by the UK Medical Research Council intramural project MC_UU_00025/3 (no. RG94521) to L.M.M. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
G.F., I.C., S.H.Y.L. and L.M.M. initiated the project. G.F., S.H.Y.L. and L.M.M. designed the study, coordinated the experiments and provided conceptual inputs for the paper. G.F., S.H.Y.L. and L.M.M. wrote the manuscript. G.F., I.C., N.S.L., S.L., A.C.C., S.H.Y.L. and L.M.M. performed experiments and analyzed data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Daewoo Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary table legends.
Supplementary Tables
The file contains Supplementary Tables 1–4.
Source data
Source Data Fig. 2
Unprocessed immunoblot for Fig. 2f.
Source Data Fig. 3
Unprocessed immunoblot for Fig. 3c.
Rights and permissions
About this article
Cite this article
Fedele, G., Loh, S.H.Y., Celardo, I. et al. Suppression of intestinal dysfunction in a Drosophila model of Parkinson’s disease is neuroprotective. Nat Aging 2, 317–331 (2022). https://doi.org/10.1038/s43587-022-00194-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-022-00194-z
This article is cited by
-
Modulating p38 MAPK signaling by proteostasis mechanisms supports tissue integrity during growth and aging
Nature Communications (2023)
-
Blocking dPerk in the intestine suppresses neurodegeneration in a Drosophila model of Parkinson’s disease
Cell Death & Disease (2023)
-
Gut mitochondrial defects drive neurodegeneration
Nature Aging (2022)