Abstract
Oestrogen receptor alpha (ERα) signalling in the ventromedial hypothalamus (VMH) contributes to energy homeostasis by modulating physical activity and thermogenesis. However, the precise neuronal populations involved remain undefined. Here, we describe six neuronal populations in the mouse VMH by using single-cell RNA transcriptomics and in situ hybridization. ERα is enriched in populations showing sex-biased expression of reprimo (Rprm), tachykinin 1 (Tac1) and prodynorphin (Pdyn). Female-biased expression of Tac1 and Rprm is patterned by ERα-dependent repression during male development, whereas male-biased expression of Pdyn is maintained by circulating testicular hormone in adulthood. Chemogenetic activation of ERα-positive VMH neurons stimulates heat generation and movement in both sexes. However, silencing Rprm gene function increases core temperature selectively in females and ectopic Rprm expression in males is associated with reduced core temperature. Together, these findings reveal a role for Rprm in temperature regulation and ERα in the masculinization of neuron populations that underlie energy expenditure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The single-cell RNA-sequencing data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE143818. The Cellprofiler pipeline used to quantify fluorescent images and source data for Fig. 2e are presented with the paper.
Code availability
Custom R scripts written for single-cell analyses and for FISH quantification, statistics and plotting are available at https://github.com/jevanveen/vanveenkammel.
References
Lovejoy, J. C., Champagne, C. M., de Jonge, L., Xie, H. & Smith, S. R. Increased visceral fat and decreased energy expenditure during the menopausal transition. Int J. Obes. (Lond.) 32, 949–958 (2008).
Slonaker, J. R. The effect of copulation, pregnancy, pseudopregnancy and lactation on the voluntary activity and food consumption of the albino rat. Am. J. Physiol. 71, 362–394 (1925).
Brobeck, J. R., Wheatland, M. & Strominger, J. L. Variations in regulation of energy exchange associated with estrus, diestrus and pseudopregnancy in rats. Endocrinology 40, 65–72 (1947).
Kopp, C., Ressel, V., Wigger, E. & Tobler, I. Influence of estrus cycle and ageing on activity patterns in two inbred mouse strains. Behav. Brain Res. 167, 165–174 (2006).
Olofsson, L. E., Pierce, A. A. & Xu, A. W. Functional requirement of AgRP and NPY neurons in ovarian cycle-dependent regulation of food intake. Proc. Natl Acad. Sci. USA 106, 15932–15937 (2009).
Sanchez-Alavez, M., Alboni, S. & Conti, B. Sex- and age-specific differences in core body temperature of C57Bl/6 mice. Age (Dordr.) 33, 89–99 (2011).
Heine, P. A., Taylor, J. A., Iwamoto, G. A., Lubahn, D. B. & Cooke, P. S. Increased adipose tissue in male and female estrogen receptor-ɑ knockout mice. Proc. Natl Acad. Sci. USA 97, 12729–12734 (2000).
Park, C. J. et al. Genetic rescue of nonclassical ERɑ signaling normalizes energy balance in obese Erɑ-null mutant mice. J. Clin. Invest. 121, 604–612 (2011).
Xu, Y. et al. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 14, 453–465 (2011).
Hulley, S. et al. Randomized trial of estrogen plus progestin for secondary prevention of coronary heart disease in postmenopausal women. JAMA 280, 605–613 (1998).
Writing Group for the Women’s Health Initiative Investigators. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the women’s health initiative randomized controlled trial. JAMA 288, 321–333 (2002).
Smith, A. W., Bosch, M. A., Wagner, E. J., Rønnekleiv, O. K. & Kelly, M. J. The membrane estrogen receptor ligand STX rapidly enhances GABAergic signaling in NPY/AgRP neurons: role in mediating the anorexigenic effects of 17β-estradiol. Am. J. Physiol. Endocrinol. Metab. 305, E632–E640 (2013).
Martinez de Morentin, P. B. et al. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab. 20, 41–53 (2014).
Herber, C. B. et al. Estrogen signaling in arcuate Kiss1 neurons suppresses a sex-dependent female circuit promoting dense strong bones. Nat. Commun. 10, 163 (2019).
Asarian, L. & Geary, N. Sex differences in the physiology of eating. Am. J. Physiol. Regul. Integr. Comp. Physiol. 305, R1215–R1267 (2013).
Rivera, H. M. & Stincic, T. L. Estradiol and the control of feeding behavior. Steroids 133, 44–52 (2018).
Musatov, S. et al. Silencing of estrogen receptor ɑ in the ventromedial nucleus of hypothalamus leads to metabolic syndrome. Proc. Natl Acad. Sci. USA 104, 2501–2506 (2007).
Correa, S. M. et al. An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Rep. 10, 62–74 (2015).
Hashikawa, K. et al. Esr1+ cells in the ventromedial hypothalamus control female aggression. Nat. Neurosci. 20, 1580 (2017).
Lin, D. et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature 470, 221–226 (2011).
Lee, H. et al. Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature 509, 627–632 (2014).
Yang, C. F. et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909 (2013).
Wang, L. et al. Hypothalamic control of conspecific self-defense. Cell Rep. 26, 1747–1758.e5 (2019).
Silva, B. A. et al. Independent hypothalamic circuits for social and predator fear. Nat. Neurosci. 16, 1731–1733 (2013).
Remedios, R. et al. Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 550, 388–392 (2017).
Narita, K., Murata, T. & Matsuoka, S. The ventromedial hypothalamus oxytocin induces locomotor behavior regulated by estrogen. Physiol. Behav. 164, 107–112 (2016).
Krause, W. C. et al. Estrogen drives melanocortin neurons to reduce sedentary behavior. Preprint at bioRxiv https://doi.org/10.1101/794792 (2019).
Flanagan-Cato, L. M. Sex differences in the neural circuit that mediates female sexual receptivity. Front. Neuroendocrinol. 32, 124–136 (2011).
Yang, T. & Shah, N. M. Molecular and neural control of sexually dimorphic social behaviors. Curr. Opin. Neurobiol. 38, 89–95 (2016).
Krause, W. C. & Ingraham, H. A. in Sex and Gender Factors Affecting Metabolic Homeostasis, Diabetes and Obesity (ed. Mauvais-Jarvis, F.) 199–213 (Springer International Publishing, 2017).
Kammel, L. G. & Correa, S. M. Selective sexual differentiation of neurone populations may contribute to sex-specific outputs of the ventromedial nucleus of the hypothalamus. J. Neuroendocrinol. 32, e12801 (2019).
Dhillon, H. et al. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 49, 191–203 (2006).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Cheung, C. C., Kurrasch, D. M., Liang, J. K. & Ingraham, H. A. Genetic labeling of steroidogenic factor-1 (SF-1) neurons in mice reveals ventromedial nucleus of the hypothalamus (VMH) circuitry beginning at neurogenesis and development of a separate non-SF-1 neuronal cluster in the ventrolateral VMH. J. Comp. Neurol. 521, 1268–1288 (2012).
Sheng, M. & Greenberg, M. E. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485 (1990).
Wu, Y. E., Pan, L., Zuo, Y., Li, X. & Hong, W. Detecting activated cell populations using single-cell RNA-seq. Neuron 96, 313–329.e6 (2017).
Van der Maaten, L. J. P. & Hinton, G. E. Visualizing high-dimensional data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Mwangi, B., Soares, J. C. & Hasan, K. M. Visualization and unsupervised predictive clustering of high-dimensional multimodal neuroimaging data. J. Neurosci. Methods 236, 19–25 (2014).
Malik, S. et al. Histone deacetylase 7 and FoxA1 in estrogen-mediated repression of RPRM. Mol. Cell. Biol. 30, 399–412 (2010).
Allison, M. B. et al. TRAP-seq defines markers for novel populations of hypothalamic and brainstem LepRb neurons. Mol. Metab. 4, 299–309 (2015).
Tannenbaum, G. S. & Bowers, C. Y. Interactions of growth hormone secretagogues and growth hormone-releasing hormone/somatostatin. Endocrine 14, 21–27 (2001).
Luo, S. X. et al. Regulation of feeding by somatostatin neurons in the tuberal nucleus. Science 361, 76 (2018).
Schick, R. R. et al. Effect of galanin on food intake in rats: involvement of lateral and ventromedial hypothalamic sites. Am. J. Physiol. Regul. Integr. Comp. Physiol. 264, R355–R361 (1993).
Scharff, A. Z. et al. Sex differences in IL-17 contribute to chronicity in male versus female urinary tract infection. JCI Insight 4, 122998 (2019).
Li, L. et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858–873.e4 (2017).
Petropoulos, S. et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026 (2016).
Herbison, A. E. Somatostatin-immunoreactive neurones in the hypothalamic ventromedial nucleus possess oestrogen receptors in the male and female rat. J. Neuroendocrinol. 6, 323–328 (1994).
Arnold, A. P. & Chen, X. What does the ‘four core genotypes’ mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol. 30, 1–9 (2009).
Kow, L.-M. & Pfaff, D. W. Estrogen effects on neuronal responsiveness to electrical and neurotransmitter stimulation: an in vitro study on the ventromedial nucleus of the hypothalamus. Brain Res. 347, 1–10 (1985).
Minami, T., Oomura, Y., Nabekura, J. & Fukuda, A. 17β-Estradiol depolarization of hypothalamic neurons is mediated by cyclic AMP. Brain Res. 519, 301–307 (1990).
Zadran, S. et al. 17-β-Estradiol increases neuronal excitability through MAP kinase-induced calpain activation. Proc. Natl Acad. Sci. USA 106, 21936 (2009).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163 (2007).
Chen, R., Wu, X., Jiang, L. & Zhang, Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017).
Romanov, R. A. et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176 (2016).
Kim, D.-W. et al. Multimodal analysis of cell types in a hypothalamic node controlling social behavior. Cell 179, 713–728.e17 (2019).
Schulz, D. J. Plasticity and stability in neuronal output via changes in intrinsic excitability: it’s what’s inside that counts. J. Exp. Biol. 209, 4821–4827 (2006).
McCarthy, M. M. & Arnold, A. P. Reframing sexual differentiation of the brain. Nat. Neurosci. 14, 677–683 (2011).
Forger, N. G., Strahan, J. A. & Castillo-Ruiz, A. Cellular and molecular mechanisms of sexual differentiation in the mammalian nervous system. Front. Neuroendocrinol. 40, 67–86 (2016).
Chartoff, E. H. & Mavrikaki, M. Sex differences in kappa opioid receptor function and their potential impact on addiction. Front. Neurosci. 9, 466 (2015).
Ferran, J. L., Puelles, L. & Rubenstein, J. L. R. Molecular codes defining rostrocaudal domains in the embryonic mouse hypothalamus. Front. Neuroanat. 9, 46 (2015).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Burgoyne, P. S. & Arnold, A. P. A primer on the use of mouse models for identifying direct sex chromosome effects that cause sex differences in non-gonadal tissues. Biol. Sex Differ. 7, 68–68 (2016).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Crane, J. D., Mottillo, E. P., Farncombe, T. H., Morrison, K. M. & Steinberg, G. R. A standardized infrared imaging technique that specifically detects UCP1-mediated thermogenesis in vivo. Mol. Metab. 3, 490–494 (2014).
Robbins, E. et al. Quantitative non-radioactive in situ hybridization of preproenkephalin mRNA with digoxigenin-labeled cRNA probes. Anat. Rec. 231, 559–562 (1991).
Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates 3rd edn (Academic Press, 2008).
McQuin, C. et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 16, e2005970 (2018).
Lein, E. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Acknowledgements
The research was supported by UCLA Division of Life Sciences funds to S.M.C., NIH grant no. K01 DK098320 to S.M.C., NIH grant no. UL1TR001881 and Iris Cantor-UCLA Women’s Health Center/UCLA National Center of Excellence in Women’s Health Pilot Awards to S.M.C. and Z.Z., UCSD/UCLA Diabetes Research Center NIH grant no. P30 DK063491 Pilot and Feasibility awards to S.M.C. and M.L., NIH grant nos. DK104363 and DK117850 to X.Y., NIH grant nos. HD076125 and HL131182 to A.P.A., UCLA Department of Medicine Chair Commitment and NIH grant no. AA026914 to M.L., predoctoral NRSA (grant no. F31 AG051381) and Hyde Fellowship to L.G.K., UCLA Dissertation Year Fellowships to L.G.K. and D.A., Canadian Diabetes Association Postdoctoral fellowship to M.S., American Heart Association Postdoctoral Fellowship (grant no. 18POST33960457) to Z.Z. and NSF Graduate Research Fellowship to M.G.M. We thank C. De La Cruz for technical assistance. This work was supported by the following facilities at UCLA: the UCLA Translational Pathology Core Laboratory, the MCDB/BSCRC Microscopy Core, the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Flow Cytometry Core Resource, and the TCGB Technology Center for Genomics and Bioinformatics (supported by grant no. P30 CA016042).
Author information
Authors and Affiliations
Contributions
J.E.V., L.G.K. and S.M.C. conceived and designed the studies. J.E.V., L.G.K., P.C.B., M.S., M.S.R., J.W.P., Z.Z., M.G.M., A.M.J., H.H. and S.M.C. acquired and analysed data. J.E.V., L.G.K., P.C.B., M.S., D.A., M.L., A.P.A., X.Y. and S.M.C. contributed to data interpretation. J.E.V., L.G.K. and S.M.C. wrote the manuscript with substantial input from M.S., Z.Z., M.G.M., D.A., M.L., A.P.A. and X.Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Elena Bellafante.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 FACS strategy for the isolation of Sf1 lineage cells.
Related to Fig. 1. a, Gating of all events using forward scatter area (FSC-A) and side scatter area (SSC-A) to select for probable cellular objects (gate p1) based on size and internal complexity. b, FSC and SSC gating to remove doublets (gate p2,p3). c, Gating using live cell permeable (Alexa 700,DRAQ5) and live cell impermeable (DAPI) DNA dyes. Objects displaying high DRAQ5 and low DAPI are nucleated and alive (gate live-1). d, Gating to select nucleated single cells displaying red fluorescence (gate p4). TdTomato+ cells were sorted into 96 well plates for downstream scRNA-Seq. e, Hierarchy of populations demonstrating that tdTomato+ (p4) cells comprise ~4% of all live, nucleated objects obtained from rough dissection and dissociation of hypothalami. f, graphs comparing FSC-A and SSC-A of live, nucleated tdTomato- objects to live, nucleated tdTomato+ objects. FACS plots are representative of two separate experiments comprised of n = 3 female mice and n = 3 male mice.
Extended Data Fig. 2 The transcriptional architecture of the VMH is similar in males and females.
Related to Fig. 2. a, tSNE showing all clusters identified by bioinformatic analyses including those predicted to be from the arcuate nucleus of the hypothalamus (Pomc) and rare non-neuronal cells (Apoe) b, tSNE showing that male and female neurons are present in all clusters identified.
Extended Data Fig. 3 Clustering and expression of broadly expressed VMH markers and markers outside of the VMH.
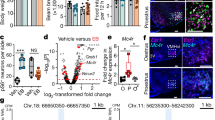
Related to Fig. 3. a, Hpcal1 expression appears diffusely in the adult male (image from Allen Brain Atlas), top, and female VMH, below (representative of images from n = 5 mice). b, Gal expression is restricted to scattered cells in the adult male (image from Allen Brain Atlas), top, and female VMH, below (representative of images from n = 1 mouse). c, expression of Tac1, Rprm, Pdyn, and Sst in the retrochiasmatic area (RCH) and ventral premammillary nucleus (PMv), adjacent to the VMH along the rostral-caudal axis in males (n = 3 mice) and females (n = 4 mice). Scalebars = 100μm, DAPI shown in cyan.
Extended Data Fig. 4 Limited overlap in expression of Sst with Tac1 or Rprm.
Related to Fig. 4. a, Sst (yellow) and Rprm (magenta) transcripts in the VMHvl (images representative of n = 5 female mice). b, Sst (yellow) and Tac1 (magenta) transcripts in the VMHvl (images representative of n = 5 female mice). c, Sst transcripts are sparse but often associated with ERα expression (images representative of n = 6 female mice). d, quantification of Sst transcripts and ERα immunoreactivity confirms that while the majority of ERα expressing cells do not co-express Sst, the majority of Sst expressing cells co-express ERα (n = 6 female mice).
Extended Data Fig. 5 Activational effects of hormones maintain male-biased expression of Pdyn.
Related to Fig. 5. a, four core genotypes of mice (n = 2 animals for all GDX panels, n = 3 animals for sham panel) analysis showing that Pdyn expression is maintained in adult males by circulating testicular hormone. b, four core genotypes analysis confirming that there is no sex difference in Sst expression(n = 2 animals for all GDX panels, n = 3 animals for sham panel). Dashed line shows boundary of VMH and VMHvl, in blue for male and magenta for female. Scalebars = 200μm.
Extended Data Fig. 6 Chemogenetic activation of Esr1+ VMHvl neurons enhances BAT thermogenesis.
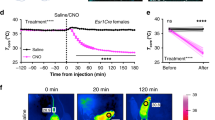
cFOS immunoreactivity in wild-type (Esr1Cre-negative, n = 2 animals) or Esr1Cre (Esr1Cre-positive, n = 3 animals) littermate female mice perfused 90 minutes after CNO injection. Scalebar = 200 μm. b, Image quantification, mean±SEM shown. c, Infrared thermography of male and female Esr1Cre mice (n = 6: 4 male mice + 2 female mice) injected with AAV-DIO-hM3Dq-mCherry, 30 minutes before (Pre-Tx) and 60 minutes after injection with CNO or saline (Post-Tx). Dashed line indicates interscapular region directly above BAT. d, quantification of shows a rise in intrascapular temperature following treatment with CNO compared to saline treatment in the same animals on a different day. Two-way RM ANOVA: pre vs post: F(1,10) = 6.331, p = .0306. Sidak’s multiple comparisons test: pre vs post CNO: t = 2.763, p = .0397; pre vs post saline: t = .7954, p = .6918.
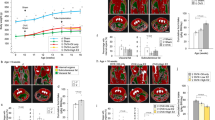
Extended Data Fig. 7 Depletion of Rprm in the VMH enhances BAT thermogenesis.
Representative thermal images of female mice injected with either Rprm targeting or non-targeting siRNA pools. b, Quantification of thermography shows a significant increase in skin temperature above the interscapular BAT depots in ovariectomized (OVX) female mice injected with Rprm targeting siRNA pools (n = 6 animals) compared to OVX female mice injected with non-targeting siRNA pools (n = 6 animals) (Two-way RM ANOVA: siRNA type (F(1,5) = 16.16, p= 0.0101); hormone treatment (F(1,5) = 0.2471, p = 0.6402); interaction (F(1,5) = 0.0005832, p = 0.9817). The effect of Rprm depletion on BAT is not changed by estrogen replacement (mean±SEM shown). c, Quantification of thermography shows no significant difference in tail skin temperature in ovariectomized (OVX) female mice injected with Rprm targeting siRNA pools (n = 6 animals) compared to OVX female mice injected with non-targeting siRNA pools (n = 6 animals) (mean±SEM shown). d, representative images of BAT histology showing a slight decrease in lipid content in female mice injected with Rprm targeting siRNA pools (n = 8 animals) as compared to female mice injected with non-targeting siRNAs (n = 8 animals). BAT was collected 14 days after siRNA injection. e, Representative images of BAT histology in male mice with developmental ablation of hypothalamic ERα (Esr1fl/fl; Nkx2-1Cre, n = 5 animals) or littermate controls (Esr1fl/fl, n = 5 animals). Scalebars = 50μm.
Supplementary information
42255_2020_189_MOESM2_ESM.xlsx
Supplementary Table 1. Complete ANOVA tables and multiple comparisons testing for all ANOVAs performed in the study. Each tab pertains to one figure panel and is named as such.
Rights and permissions
About this article
Cite this article
van Veen, J.E., Kammel, L.G., Bunda, P.C. et al. Hypothalamic oestrogen receptor alpha establishes a sexually dimorphic regulatory node of energy expenditure. Nat Metab 2, 351–363 (2020). https://doi.org/10.1038/s42255-020-0189-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0189-6
This article is cited by
-
CaMKIIa Neurons of the Ventromedial Hypothalamus Mediate Wakefulness and Anxiety-like Behavior
Neurochemical Research (2023)
-
Populational heterogeneity and partial migratory origin of the ventromedial hypothalamic nucleus: genoarchitectonic analysis in the mouse
Brain Structure and Function (2023)
-
CerS6-dependent ceramide synthesis in hypothalamic neurons promotes ER/mitochondrial stress and impairs glucose homeostasis in obese mice
Nature Communications (2023)
-
Transcriptome-scale spatial gene expression in rat arcuate nucleus during puberty
Cell & Bioscience (2022)
-
The ventromedial hypothalamic nucleus: watchdog of whole-body glucose homeostasis
Cell & Bioscience (2022)