Abstract
To identify candidate disease genes of central nervous system (CNS) phenotypes, we created the Neurogenetic Systematic Correlation of Omics-Related Evidence (NeuroSCORE). We identified five genome-wide metrics highly associated with CNS phenotypes to score 19,601 protein-coding genes. Genes scored one point per metric (range: 0–5), identifying 8298 scored genes (scores ≥ 1) and 1601 “high scoring” genes (scores ≥ 3). Using logistic regression, we determined the odds ratio that genes with a NeuroSCORE from 1 to 5 would be associated with known CNS-related phenotypes compared to genes that scored zero. We tested NeuroSCORE using microarray copy number variants (CNVs) in case–control cohorts and aggregate mouse model data. High scoring genes are associated with CNS phenotypes (OR = 5.5, p < 2e-16), enriched in case CNVs, and mouse ortholog genes that cause behavioral and nervous system abnormalities. We identified 1058 high scoring genes with no disease association in OMIM. Transforming the logistic regression results indicates high scoring genes have an 84–92% chance of being associated with a CNS phenotype. Top scoring genes include GRIA1, MAP4K4, SF1, TNPO2, and ZSWIM8. Finally, we interrogated CNVs in the Clinical Genome Resource, finding the majority of clinically significant CNVs contain high scoring genes. These findings can direct future research and improve molecular diagnostics.
Similar content being viewed by others
Introduction
A cholera epidemic swept across the globe in 1819 from India through the Middle East, Europe, and to America. British- and American-based physicians John Snow and Amariah Brigham both studied cholera and produced maps of the deaths in New York and London—Brigham’s in 1831 and Snow’s in 18551,2. Both maps used different overlapping evidence, such as trade routes and drinking water systems, to illustrate a confluence of variables leading to new insights about cholera and, ultimately, public health remedies. Today, geneticists can take a similar approach, with different types of maps, to identify the genetic mechanism of diseases. With genome-wide, multi-omic analyses we can now overlay these datasets on the human genome and correlate these with phenotypes of the central nervous system (CNS) to identify candidate disease genes thereby improving research into candidate disease genes, diagnostics and, eventually, therapies.
Identifying disease or risk genes for conditions of the CNS has been a slow process, with current diagnostic rates for children with a broad range of neurological or developmental conditions ranging from 31% to 53%3,4 (undergoing multiple clinical tests) and approximately 32% in adults5. Diagnostic rates using whole exome sequencing and chromosomal microarray vary within particular phenotypes, ranging from approximately 16% in autism spectrum disorder (ASD)6, 23% in corpus callosum anomalies7, and 42% in early-onset epileptic encephalopathies8. These conditions all likely have substantial unrecognized genetic contribution and a recent study of developmental disorders found that more than 1000 additional genes are expected to contribute, either alone or in combination, to neurodevelopmental disorders9.
Identifying CNS-disease genes is complicated, as genetic CNS diseases are caused by multiple pathogenic mechanisms4, display multiple forms of inheritance, are characterized by allelic heterogeneity, reduced penetrance, pre/perinatal lethality, and variable expressivity, are difficult to study in vivo, have phenotypes that exist on a spectrum (e.g. ASD), have variable age of onset, have broad descriptions (e.g. “developmental delay”), and many genes characterized in the 1980s–1990s have received disproportionate study leading to many unstudied genes, colloquially called the “ignorome”10.
Previous attempts to identify candidate disease genes have used multiple approaches, including statistical modeling for probability of causing a haploinsufficiency-related condition (pLI score11) or identifying genes with regional coding constraint12. Other systems rely on direct counts of particular type of variant observed in a control population (gnomAD’s observed/expected metrics13). Lastly, systems have been devised to search for disease-specific genes, such as ForecASD14 and candidate ASD genes, or used specific data such as gene expression patterns in the brain-coX model15. These systems, while useful, are limited and no multi-omic system has yet been devised for CNS phenotypes.
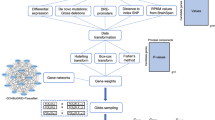
As a diagnostic laboratory focused on neurological and developmental phenotypes, we sought to create a model that identified and prioritized candidate disease genes. We began with two foundational concepts, the first being the use of a multi-omics approach to account for different potential disease mechanisms and characteristics of genes that underlie known neurogenetic conditions. The second concept is developmental brain dysfunction, which posits that distinct clinical diagnoses affecting the cognitive, motor, neuropsychiatric, neurobehavioral, or neuroanatomical domains can be classified into an overarching umbrella term that describes the underlying cause of clinical symptoms because they share similar underlying genetic risk factors16,17. By leveraging large scale -omics databases supporting these two foundational concepts with a clinical database, we created NeuroSCORE: the Neurogenic Systematic Correlation of Omics-Related Evidence (Fig. 1). We believe NeuroSCORE is the first multi-omic model to assess most protein-coding genes and focused broadly on CNS phenotypes.
Schematic of final NeuroSCORE model. Key: De novo: De Novo Database; gnomAD: Genome Aggregation Database; Critical Constraint: Critically constrained coding regions database; GTEx: Gene-Tissue Expression database; Index: Database of genes based on Uddin et al.66; OMIM: Online Mendelian Inheritance in Man database (see “Methods”).
Results
Constructing and assessing the NeuroSCORE model
To build a comprehensive and clinically useful model, we chose genome-wide databases with gene-specific data to assess expression, constraint, and other properties of protein coding genes (dependent variables; see “Methods”). Using Pearson’s chi squared, we determined if any of the identified nine gene metrics were correlated with our outcome measure of genes currently associated with a condition that has one or more CNS-related phenotypes in the OMIM database. Briefly, these gene metrics identify genes with: significant enrichment of de novo variants in individuals affected with neurological or neurodevelopmental conditions (“de novo” metric), one or more exons with high expression throughout human brain development and with low mutation burdens (“Index” metric), one or more regions of coding constraint at the 95th or 99th percentiles as measured in samples from the Genome Aggregation Consortium (gnomAD) called “critically constrained regions” (“CCR 99” and “CCR 95” metrics), median brain expression at or above 10 transcripts per million as measured across all brain region samples from the Genotype-Tissue Expression (GTEx) database (GTEx metric), low levels of loss-of-function or missense variants in gnomAD (“gnomAD LOF” and “gnomAD MIS” metrics), and enrichment in copy number variants (CNVs) in a neurodevelopmentally affected cohort at different levels of statistical significance (“Coe 1” and “Coe 2” metrics).
Eight of these nine variables were significantly associated with currently known CNS-related disease genes (Table 1), including the Genotype-Tissue Expression (GTEx) database genes, de novo genes, genes with regions of critical constraint at both the 99th and 95th percentiles, gnomAD LOF and MIS genes, Index genes, and Coe 1 genes metrics (all p < 0.05). The Coe 2 gene metric was not correlated (p = 0.10), and data were excluded. Using the remaining eight significant metrics, we then constructed a multiple logistic regression model. Main effects from multiple logistic regression modeling indicated that five of the eight metrics were significantly and positively associated with odds ratios (ORs) above 1.0 for the outcome measure of CNS-related disease genes.
These five variables became our final NeuroSCORE model: de novo genes, Index genes, critically constrained genes at the 99th percentile, GTEx genes, and gnomAD LOF genes. Two-way interactions were not assessed; however, all ORs are within a similar range suggesting these variables have a similar strength and thus no single variable is having an outsized influence on the model.
NeuroSCORE creates different scoring levels for genes from 0 to 5 points. We then used logistic regression to investigate the relative enrichment of currently known CNS-related disease genes within each scoring level compared to genes that scored zero and calculated the OR for genes identified at each scoring level (Table 2 and Fig. 2). ORs increased with each increase in NeuroSCORE, ranging from 2.0 to 32.2 (p < 5e-8). Next, we calculated the OR for genes that met a majority of the NeuroSCORE metrics (≥ 3 points), which we considered to be “high scoring” genes (Table 2). As this set of high scoring genes is significantly associated with CNS-related disease genes, we focused the remaining analyses on the high scoring genes (N = 1601). As a negative control, we calculated the OR for genes that scored 0 in our model to be associated with non-CNS-related phenotypes. This set of genes was significantly more likely to be associated with genes causing non-CNS-related phenotypes (OR = 1.7, 95% CI 1.5–1.9, p < 2e-16), showing these genes are enriched for non-CNS-related conditions.
High scoring genes and mouse model organism data
To further assess our model, we applied NeuroSCORE to the 27 high-level phenotypes of mouse model data curated by Jackson Labs Mouse Genome Informatics database18. Among genes with high-level experimental phenotype data (N = 8149), total phenotypes per gene ranged from 1 to 26 with an average of 5.99 phenotypes (SD = 4.55). Using chi squared analysis between high scoring gene orthologs and the presence of any of the 27 phenotypes (Bonferroni corrected for 27 tests, p < 0.002), we found high scoring gene orthologs were significantly enriched in seven phenotypes including: mortality and aging, embryonic abnormalities, central and peripheral nervous system abnormalities, growth and congenital anomalies, abnormalities of learning and behavior, abnormalities of cellular proliferation, differentiation, and apoptosis, and abnormal muscle development (all p ≤ 2.9e-5).
Using logistic regression, we found high scoring genes were significantly more likely to be associated with mouse ortholog genes that cause the behavioral phenotypes (OR = 1.5, 95% CI 1.3–1.7, p = 5.9e-10) and central/peripheral nervous system phenotypes (OR = 1.7, 95% CI 1.5–1.9, p = 4.7e-16), while they were significantly depleted in genes causing non-neurological phenotypes (OR = 0.64, 95% CI 0.57–0.73, p = 3.8e-12; see Fig. 3D).
GO enrichment and pathway analyses
We performed GO Analyses on the set of 1601 high scoring genes to determine if enrichment of key biological processes, cellular components, and molecular functions occurred within this set. Enriched terms in biological processes include positive regulation of protein localization to Cajal body (GO:1904871), axo-dendritic protein transport (GO:0099640), and alternative mRNA splicing, via spliceosome (GO:0000380). Within cellular components, key terms included the nBAF complex (GO:0071565) and the NuRD complex (GO:0016581), which are involved in chromatin remodeling. For the molecular functioning area, terms included binding activity such as protein kinase A catalytic subunit binding (GO:0034236), microtubule plus-end binding (GO:0051010), and pre-mRNA binding (GO:0036002). See Supplemental Table S1 for a list of all of the top five enriched, unique GO annotation terms and high scoring genes for each of the three areas.
From our pathway analyses, we analyzed approximately 500 GO terms with the lowest false discovery rate values (all q-values ≤ 1.3e-22) by inspecting the relationships using the AmiGO visualization tool (accessed December 10th, 2020). Within the developmental pathways, the following were enriched: regulation of dendrite development (GO:0050773), regulation of morphogenesis involved in differentiation (GO:0010769), positive regulation of neurogenesis (GO:0050769), and positive regulation of neuron projection development (GO:0010976). Within the metabolic and enzymatic pathways, the following were enriched: regulation of mRNA stability (GO:0043488), regulation of mRNA splicing via the spliceosome (GO:0048024), and catalytic step two of the spliceosome (GO:0071013). Top selected terms and associated genes are presented in Supplemental Table S2. The GO and pathway analyses further support that NeuroSCORE identifies genes in important neurological and developmental processes.
Enrichment of NeuroSCORE genes in brain-expressed genes
Assuming that genes with elevated brain expression are biologically important for brain growth, development, and/or functioning, we assessed the distribution of genes identified by NeuroSCORE in the Human Protein Atlas. Using chi squared tests, we foudn that NeuroSCORE genes represent a significantly higher proportion of the genes with elevated brain expression for both high scoring and any scoring genes. High scoring genes represent 12.3% (301 of 2442) of genes with elevated expression, compared to 8.5% (1601 of 18,759) for all genes in the genome (χ = 30.5, p = 3e-8). Comparing genes that received any NeuroSCORE (scores 1–5), we similarly find that genes with elevated brain expression represent 55.3% (1351 of 2442) compared to 44.2% (8298 of 18,759) for all scored genes in the genome (χ = 37.7, p = 8e-10). This provides additional evidence that NeuroSCORE identified genes relevant to biological function (and likely dysfunction) of the CNS.
Case–control analyses in neurodevelopmentally affected and typically developing cohorts
After identifying increased ORs for high scoring genes in CNS phenotypes and showing further support for the model through mouse, GO, and brain expression analyses, we next investigated the NeuroSCORE content of human copy number variants (CNV) data. We applied NeuroSCORE to CNV data derived from microarray testing of individuals in both a population control19 and a neurodevelopmentally affected cohort20. We compared average CNV size, total genes within CNVs, total number of genes identified by NeuroSCORE in the CNVs, and median and average NueroSCORE of the CNV (p-values used Bonferroni correction of p < 0.005 corresponding to five statistical tests for two classes of CNVs). For easier interpretation of some of the following analyses, NeuroSCOREs were converted from total points to a percentage of total points (0 = 0%, 1 = 20%, 2 = 40%, 3 = 60%, 4 = 80%, and 5 = 100%).
Using two-sided t-tests, we found the average size of both loss and gain CNVs was significantly larger in cases than controls (losses: 344 kilobases (kb) vs. 104 kb, p = 3e-8; gains: 419 kb vs. 244 kb, p = 5e-8). Within gain CNVs, the average number of genes was similar between cases and controls (4.1 v. 3.6, p = 0.1), while there were slightly more genes on average within loss CNVs for cases versus controls (3.2 vs. 2.4, p = 0.001). The small range in the average number of genes in case and control CNVs (2.4–4.1 genes) suggests specific genes within case CNVs, rather than the total number of genes, could drive the neurodevelopmental phenotypes. Therefore, we investigated the NeuroSCORE profile of these CNVs to assess for differences between cases and controls.
To assess potential differences in gene content, we next analyzed median and average CNV NeuroSCORE and the distribution of scored genes between case and control CNVs. Using two-sided t-tests, we found that case CNVs had higher median and average NeuroSCORE (median: losses: 13.4% vs. 6.7%, p = 2e-16; gains: 14.9% vs. 8.5%, p = 2e-16; average: losses: 14.2% vs 7.6%, p = 2e-16; gains: 16.4% vs. 10.5%, p = 2e-16). To assess the distribution of scored genes within CNVs, we used chi squared analyses using the highest scoring gene in a CNV at each different scoring levels (5, 4, 3, 2, 1, 0, and no score/NA). Using a Bonferroni corrected p-value of p < 0.004 for 14 tests (7 scoring levels, two classes of CNVs), we found that case CNVs were significantly enriched for the high scoring NeuroSCORE genes in both loss and gain CNVs while controls were enriched for genes that achieved no score (0) in loss CNVs or were not scored/NA in both loss and gain CNVs (Table 3 and Fig. 3A, B). In case CNVs, 16.2% of losses and 21.8% of gains contained at least one high scoring gene compared to 0.6% and 4.2% of controls, respectively. Logistic regression showed a significantly increased OR for case CNVs containing one or more high scoring genes compared to controls (OR = 9.3, 95% CI 7.4–11.8, p = 2e-16). Taken together, these results support the ability of NeuroSCORE to identify differences in CNVs associated with neurodevelopmental features compared to population controls.
We next performed sub-analyses of case–control CNV by gender (NMALES = 1724, NFEMALES = 468), inheritance (NPATERNAL = 35, NMATERNAL = 43, Nde novo = 33), and clinical classification. Classifications included common variants (“population”) and those classified as likely benign, variants of uncertain clinical significance (VOUS), and clinically significant (NPOPULATION = 2195, NLIKELY BENIGN = 39, NVOUS = 148, NCLIN. SIG = 96; Fig. 3C).
Previous work has shown that the gender bias in neurodevelopmental conditions is partly due to the burden of CNVs or sequence variants21. Given this, we questioned if affected females had CNVs with higher NeuroSCOREs than affected males. Analyzing case CNVs by gender showed no differences for average NeuroSCORE (15.1% vs. 15.7%), median NeuroSCORE (13.7% vs. 14.6%), or rates of high scoring genes within CNVs (0.4 vs. 0.3). Similarly, we did not find differences in CNVs by inheritance (inherited vs. de novo) for average NeuroSCORE (20.6% vs. 26.1%), median NeuroSCORE (17.3% vs. 25.2%), or rates of high scoring genes (1.8 vs. 0.7). However, using linear regression and controlling for CNV size we found that CNVs with increasing pathogenicity showed an increase in the total number of high scoring genes (β = 0.5; p = 2e-16) and lower scoring genes (β = 0.02; p = 0.02), while non-scored genes decreased slightly (β = −0.05; p = 2.5e-8, Fig. 3C). As expected, linear regression also showed median CNV NeuroSCORE increased with increasing classification as well (β = 6.7%; p < 2 e-16). These data show high scoring genes appear to be an important component in CNVs identified in this clinically affected cohort.
Landscape of high scoring genes in pathogenic CNVs
Given pathogenic CNVs underlie a significant proportion of many neurological disorders, we applied NeuroSCORE to a set of common pathogenic CNVs as described in > 10,000 individuals with neurological or neurodevelopmental phenotypes22. All CNVs had at least one scored gene and all but the 1q21.1 distal deletion CNV had at least one high scoring gene (Fig. 4).
We next applied NeuroSCORE to the 72 CNV regions with a completed haploinsufficiency and triplosensitivity review in ClinGen, the Clinical Genome Resource which provides expert curation of the clinical relevance of genes and genetic variants (accessed January 20, 2021, Supplemental Table S3). There are currently 38 regions designated by ClinGen as having sufficient evidence for haploinsufficiency and 21 regions designated with sufficient evidence for triplosensitivity. Although some regions may vary in size, we used the coordinates provided by ClinGen (hg19/GRCh37). The majority of these regions contain at least one gene with a high NeuroSCORE (30/38 and 14/21, respectively). Next, we explored the NeuroSCORE profile of several recurrent CNV regions to help identify genes that are likely to contribute to CNS-related phenotypic features.
The 7q11.23 recurrent region is approximately 1.5 megabases with deletions associated with Williams-Beuren syndrome (WBS) and gains associated with 7q11.23 duplication syndrome. The typical WBS deletion/duplication region contains 25 total genes, of which 15 are scored and six are high scoring genes. The GTF2I and GTF2IRD1 genes have been implicated as key genes driving the neurobehavioral phenotype23,24, though GTF2I and GTF2IRD1 knockout mice suggests that neither gene fully recapitulates the neurobehavioral aspects of WBS23. Aside from GTF2I, NeuroSCORE identified five high scoring genes (Fig. 4), all with evidence for CNS involvement: STX1A has been associated with ASD25 and syndromic ID26 in humans, LIMK1 sequence variants are associated with ASD27 and visuospatial impairment28, while LIMK1 deficient mice have fewer cortical pyramidal neurons29; BAZ1B knockout mice show abnormal neurogenesis30 while clinical studies show variants associated with ASD27, Klippel-Feil syndrome31, and congenital heart defects27; EIF4H-deficient mice have a smaller body size, behavioral impairments, and reduced brain volume32; finally, a single CLIP2 variant has been associated with ASD33.
The 22q11.2 region is also associated with recurrent deletion and duplication syndromes. Within the typical 22q11.2 region there are 64 genes, of which 38 are scored and eight are high scoring genes. However, the 22q11.2 deletion/duplication syndrome region presents a challenge when interpreting CNVs that are smaller than the common breakpoint in the A-F deletion (breakpoints refer to areas of repetitive DNA segments that cause recurrent CNVs and often are given letter designations). Our analysis found the 22q11.2 A–B, B–D, and D–F breakpoint CNVs each contain multiple scored genes and at least one high scoring gene (Fig. 4), suggesting that CNVs of any of these smaller regions of 22q11.2 may be pathogenic for CNS-related clinical features. Two previous studies in cohorts of individuals with 22q11.2 deletion syndrome are also consistent with our data. The first study analyzed neuroimaging and transcriptomic data and identified the MAPK1 gene (a gene in the D-F region with NeuroSCORE of 4) as a potential driver gene of brain morphology changes34. The second study analyzed 22q11.2 deletion size and IQ score, finding that IQ score was partially explained by deletion size (A–B vs. B–D)35. Taken together, NeuroSCORE identified several candidate genes in the 7q11.23 and 22q11.2 regions, some of which are supported by other studies as well as additional genes that may provide new insight into the role of these regions in neurological phenotypes.
Emerging CNV syndromes also pose challenges for clinical interpretation. Applying NeuroSCORE to rare CNVs can help determine if they score in the same range as known pathogenic CNVs while also identifying candidate genes. We queried the ClinGen Dosage Sensitivity Map and identified the 2p16.1-p15 CNV (chr2:54,700,000–63,900,000; hg19) as one with limited data on whether it caused a duplication syndrome. To date, seven individuals have been reported with a duplication and neurological clinical features36,37,38. Experimental studies in zebrafish of the previous candidate genes have implicated the BCL11A, USP34, REL, and XPO1 genes39 as possibly associated with a phenotype. Using NeuroSCORE, we first find this CNV has a median score of 30%, well above the average median score of 16% in gain CNVs in our affected case cohort. Furthermore, within this region are four high scoring genes (BCL11A, USP34, XPO1, and CCT4), of which the CCT4 gene was not identified by previous studies. Experimental work in Drosophila shows CCT4 knockdown results in severely reduced dendritic growth40; furthermore, two de novo, missense variants have been reported in two individuals with ASD27. These data support a role for 2p15-p16.1 gains in CNS phenotypes and indicate the CCT4 gene as a new candidate gene.
Discussion
We hypothesized that CNS-related disease genes are likely to be identified by multiple metrics that assess different genic properties. After combining multiple genome-wide databases that assess different properties, we found that five metrics were independently and positively associated with genes already known to cause CNS-related conditions as identified in the OMIM database. These five metrics have similar odds ratios in logistic regression modeling (range: 1.4–2.2) and suggest that they have similar strengths of association with known neurogenetic disease genes and that no single variable has an outsized influence on the model. A total of 8298 genes were identified by at least one of five gene metrics and 1601 genes were identified by three or more. These high scoring genes more often have elevated gene expression in the human brain, are enriched in developmental and neurological pathways, show neurologically related phenotypes in murine experiments, and are affected by CNVs more often in a neurodevelopmentally-affected cohort (Table 3 and Fig. 3). Conversely, genes that scored 0 in our model were enriched for genes that are associated with non-CNS-related phenotypes. Of these 1601 high scoring genes NeuroSCORE identified, 1058 (66%) do not yet have any phenotype association in OMIM (accessed July 31st, 2021) and 210 genes (13%) have no associated variants in the Human Genome Mutation Database (HGMD; accessed November 11th, 2021).
When we transform the logistic regression results of high scoring genes to the logistic likelihood curve, we can determine the likelihood that any gene within the set will cause a condition with CNS-related features. Transforming these results shows that genes within this set have an 84–92% chance of being associated a condition that results in CNS-related features, suggesting the substantial majority of the 1601 genes are likely to eventually be associated with neurogenic disorders. These 1601 gene likely represent a proportion of undiscovered neurodevelopmental genes proposed by recent analysis9. However, NeuroSCORE also captures genes related to neuropsychiatric or neurodegenerative disease. Analyses of genes associated with phenotypes such as ataxia, Parkinson’s, and amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD) showed the majority of genes are scored (ataxia: N = 4 of 5; Parkinson’s: N = 19 of 21, and ALS/FTD: N = 26 of 32). Further, many fall into the high scoring range (ataxia: N = 2 of 5; Parkinson’s: N = 7 of 21, and ALS/FTD: N = 8 of 32). Building on the NeuroSCORE framework to target more specific phenotypes may further broaden its applicability in the future.
The findings from our pathway and GO analyses found significant enrichment in multiple neurological processes, many with known disease or phenotype associations (Supplemental Tables S1 and S2): the BAF (or SWI/SNF) complex41, the NuRD complex42, neuronal organization with microtubule tracking43, tau protein activity44, histone binding processes45, the Cajal body46,47, proteasome activity48, mRNA processing via both the spliceosome components49 and mRNA trafficking and binding50. Of these processes, splicing may be one of the most important. Tissue-specific splicing in the brain has shown high rates of alternative transcript splicing, suggesting that splicing proteins and proper splicing are imperative to neuronal development, structure, and function and appear to be evolutionarily conserved51. The Cajal body presents an interesting confluence of multiple previously discussed CNS-disease related processes as these nuclear bodies appear in fetal and neural cells to help mediate splicing, create parts of the spliceosome and ribonuclear proteins, activate transcription, and aid in chromatin and genome organization46. Considering that ASD and other neurodevelopmental disorders appear to begin in the fetal period52, the enrichment of Cajal body-associated genes in our analyses raises an interesting target for additional study of genes related to possible “Cajalopathies”.
Our model identified 123 high scoring genes that are associated with conditions in OMIM that have no known CNS-related clinical features (OMIM accessed July 31st, 2021). While some of these genes are likely false positives and will not be found to cause CNS-related features, some have emerging evidence for causing CNS-related phenotypes. One example is MORC2, currently associated with only a form of Charcot-Marie-Tooth disease (OMIM #616661) but recently reported to cause a neurodevelopmental disorder53. Using NeuroSCORE could help identify CNS disease genes that have been overlooked due to prior disease associations (see Supplementary Table S4 for a list of genes).
Finally, our model may be helpful to identify genes that influence or increase the risk for spectrum conditions, such as ASD. Multiple damaging variants in multiple scored genes could explain the risk or presence of a condition like ASD in individuals without a known variant in high-risk genes or common pathogenic CNVs. Damaging variants in scored genes may also help explain variable expressivity and reduced penetrance observed in many CNS-related genetic conditions (e.g., “two-hit” models).
Like Amirah Brigham’s and John Snow’s use of mapping in the nineteenth century cholera epidemic, we have correlated existing data and created a new map to aid in discovery of conditions that broadly effect humanity. Our NeuroSCORE map of the human genome identifies and prioritizes potential disease genes of the CNS, which we validated using case–control and mouse model organism data. NeuroSCORE can be used for bioinformatic analysis pipelines, identification of candidate disease genes in individuals with neurological phenotypes, guidance of basic and clinical research, the development of genetic tests, and furthering research on treatments for these conditions as current or future medications may target specific proteins or pathways. Future directions of model development can include identifying interaction terms to improve model precision as well as adding new metrics such as transcriptome or methylation profiles from brain expressed genes. While there are genes that cause CNS-related conditions not identified by NeuroSCORE (e.g., GABRG2), our model represents a potentially significant step forward in research to improve ultimately diagnostics for individuals with genetic causes of neurological conditions.
Limitations
One limitation to this study and model is that it analyzes only protein-coding genes and excludes disease mechanisms such as mitochondrial, epigenetic, and disruption of enhancer and untranslated regions. Recent work in a small ASD cohort indicates that certain neurological conditions, such as ASD risk, may be influenced by variants in non-coding regions54. Similarly, genes causing autosomal recessive conditions are not well represented due to the use of gnomAD loss-of-function data. However, a recent analysis in the Deciphering Developmental Disorders cohort found approximately 3.6% of individuals from non-consanguineous families had a condition consistent with a recessive inheritance pattern55. Phenotypes caused by a significant proportion of environmental factors or oligogenic risks are similarly not well identified using NeuroSCORE. Another limitation is that our outcome variable (CNS clinical features) is drawn from OMIM, which is a rigorously maintained database but also an imperfect store of genotype–phenotype information due to possibly inaccurate or outdated information and ascertainment bias. Finally, many conditions are not yet fully phenotyped, with rare phenotypes or age-related phenotypes not well represented.
Methods
Building the NeuroSCORE model
To build a comprehensive and clinically useful model, we chose genome-wide datasets with gene-specific data and combined them. As this analysis focused on protein-coding genes, we excluded non-coding genes, RNA-coding genes, genes in the mitochondrial DNA, and pseudogenes. We sought lines of evidence previously associated with neurodevelopmental or neurological phenotypes including loss of function constraint11,13, constrained coding regions12, de novo variation27,56,57,58,59,60, brain expression levels61, copy number variation58,62, and genes with exons that are both highly expressed in brain tissues and under purifying selection63. If a gene was identified by one of the following metrics, it received a score of one point (a categorical variable, yes vs. no). We began with seven preliminary gene metrics (possible independent variables) from which to build our model. Of note, two of these metrics have two levels—one more restrictive than the other—yielding nine total possible variables from which to begin exploratory analyses. Of these nine, only five were retained in our final model, as described in the Results (Fig. 1) and designated by an asterisk (*) below:
-
1.
gnomAD LOF*: The gene’s upper bound score of the gnomAD observed/expected (o/e) loss-of-function metric was < 0.35, the preferred cutoff stated on the gnomAD site13, with rounding from the thousandths place (i.e., genes below 0.345). Using gnomAD v2.1 (accessed May 2019), there were 2896 genes identified by the gnomAD LOF gene metric.
-
2.
Critical Constraint*: The gene contained at least one area of regional constraint (critically constrained regions; CCRs) at or above the 95th or 99th percentile as described by Havrilla et al.12. The CCR 95 and CCR 99 gene metrics identified 7049 and 1444 genes, respectively, and were chosen as cutoffs as suggested in the original paper. Only genes identified at the 99th percentile were included in the final model.
-
3.
GTEx*: The median brain expression across the 13 brain tissues assessed by the Gene-Tissue Expression database v8 was ≥ 10 transcripts per million (“GTEx genes”)64. This cutoff was chosen based on the recommendation of the European Bioinformatics Institute (https://www.ebi.ac.uk/gxa/FAQ.html). The GTEx gene metric identified 6069 genes.
-
4.
De novo*: The gene was enriched for de novo variants as reported in the de novo Database using the non-Simon Simplex Cohort data (assessed January 17th, 2020)65. Regardless of potential variant pathogenicity, variants within protein-coding genes or the 3’ or 5’ untranslated regions from 13,168 trio or quartet exome/genome probands were collated from 31 unique studies for the following phenotypes: epilepsy, ASD, developmental delays, cerebral palsy, bipolar disorders 1 and 2, schizophrenia, early-onset Alzheimer and Parkinson disease, intellectual disability, neural tube defects, sporadic infantile spasm syndrome, and Tourette syndrome (see Supplementary Table S5 for a description of all studies used). We adopted a conservative approach to define a gene enriched with de novo variants if the genes contained ≥ 10 reported de novo variants. The de novo gene metric identified 487 genes.
-
5.
Index*: The gene was identified by a previous exon indexing tool (“Index genes”) with exons expressed at or above the 90th percentile in 388 post-mortem brain samples and below the 10th percentile in mutational burden for rare (< 5%) missense or loss-of-function variants in the 1000 genomes database63,66. Although specific exons within a gene are identified with this tool, we scored the entire gene if ≥ 1 exon in the gene was identified. (Note: The cutoffs differ from those originally reported in the Uddin et al.63 paper as they are more stringent and are used by Lineagen, Inc. in interpretation of clinical testing.) The Index gene metric identified 4646 genes.
-
6.
gnomAD MIS: The gene’s upper bound score of the gnomAD o/e missense variant metric was < 0.35 (the preferred cutoff stated on the gnomAD site)13. The gnomAD MIS gene metric identified 112 genes. Ultimately, this gene set was not used in our model (see “Results”).
-
7.
Coe et al.: The gene was enriched in the Coe et al.62 case–control study of individuals with childhood developmental conditions and CNVs. This study was used as it has gene-level statistics on enrichment in a neurodevelopmentally affected cohort. We used two cutoff points for the Coe gene metrics based on two significance values: p ≤ 0.01 (Coe 1 = 3116 genes) or p ≤ 0.02 (Coe 2 = 3732 genes). These p-value cutoffs were used historically in our clinical testing to determine genes or regions of potential clinical relevance. Ultimately, Coe et al. data were not used in our final model (see “Results”).
Our outcome (independent) variable was defined as whether or not a gene was associated with a phenotype containing one or more CNS-related clinical features in OMIM; this was also categorical variable (1 vs. 0, yes vs. no) and was assessed regardless of mode of inheritance. We used a previous definition of the CNS as including only the brain and spinal cord67. While an exhaustive list of CNS phenotypes meeting inclusion criteria are not possible here, examples include developmental or psychomotor delays, specific delays (e.g., speech), developmental regression, intellectual disability (or “mental retardation”), ASD or autistic traits, para- or diplegias, seizures or epilepsy, EEG abnormalities, structural anomalies of the brain or spinal cord, hydrocephalus, altered pain tolerance, sleep abnormalities, movement or coordination disorders, ataxia, tone abnormaltieis, behavioral abnormalities (e.g., aggression), psychiatric disorders, hallucinations, personality changes, emotional lability, hyper- or areflexia, Parkinson’s disease, Alzheimer’s disease, frontotemporal dementia, amyotrophic lateral sclerosis, and others. We excluded phenotypes affecting the eye, retina, cochlea, and peripheral nervous system, or conditions that caused CNS involvement due to an external event (e.g., thromboembolism). Metabolic conditions and mitochondrial conditions caused by nuclear genes were included as having CNS phenotypes as the cellular dysfunction leading to symptoms originates within the cells of the CNS. Although the retina is derived from the CNS, phenotypes involving only the retina were excluded as these conditions are treated clinically as ophthalmological conditions. Two authors (KD and MR) reviewed the top 13,021 genes in OMIM (ranked by median brain expression) which included 1822 genes with a phenotype including CNS-related clinical features and 1513 genes with phenotypes that did not have CNS-related clinical features.
Merging databases and data fidelity
Due to genes having multiple historic names, we matched data between databases by both gene name and Ensemble ID. One author (KD) visually inspected all genes identified by the primary data sources and cross-referenced discrepant or missing data with external databases (e.g., HUGO) to ensure data was present. If a gene name was discrepant, the name was updated to the current HGNC-approved name. In total, our model assesses 19,601 genes, while data for 842 genes was not available from at least one gene metric; these genes were scored as “NA”.
Statistical analyses of the NeuroSCORE model
We first assessed each of the seven gene metrics and their association with genes currently known to cause or contribute to CNS-related neurological phenotypes. We initially performed Pearson’s chi squared on each gene metric then included the variables significant at p < 0.05 in a multiple logistic regression model. Nine total variables were assessed (seven metrics with the Coe and CCR gene metrics having two levels). Using SAS v9.4, we constructed a multiple logistic regression model with backward elimination to remove variables with high multicollinearity or those that were not associated with CNS-related clinical features at p < 0.05. Wald testing was used to determine if each of the variables in the final model were significantly different from zero.
Using multiple logistic regression, we examined main effects and determined the odds ratios (ORs) for the final five metrics to be associated with genes known to cause CNS-related clinical features. We measured ORs for each variable in the final NeuroSCORE model as well as genes identified by multiple metrics (NeuroSCOREs 2–5). Both analyses used a comparison group of 4723 genes that were not identified by any metric (NeuroSCORE of 0). This yielded 1133 genes with a score of 0 that were linked to any known phenotype in OMIM (through June 2020). We performed a power analysis for these genes with a NeuroSCORE of 1 to determine the minimal detectable OR given our sample size. Setting β = 0.95 and α = 0.05 for this group of genes, the minimum OR we could detect was 1.4. All odds ratios were calculated using R using v.1.2.1335 or SAS v9.4; power analyses were performed in R with the EpiR package v2.0.17.
Evaluation of NeuroSCORE model in real-world case and control cohorts
We used two published cohorts to evaluate NeuroSCORE. As exome analyses are often performed with priority to genes already known to be involved in genetic conditions, we used copy number variant (CNVs) from microarray data from individuals affected with neurological conditions and population control cohorts. This is because CNVs often contain multiple genes of known and unknown function and significance. We matched all genes in all included CNVs to their corresponding NeuroSCORE by gene name and visually inspected and corrected all discrepancies. CNVs with only non-scored genes (e.g., pseudogenes) were omitted from analysis.
The population control cohort was drawn from the Ontario Population Genomics Platform reporting on CNVs from 1000 adults, providing 6965 total CNVs19. After removing CNVs that did not affect at least one exon of one gene, the control cohort contained 1862 gain CNVs and 2547 loss CNVs. For the case comparison group, we began with a previously published cohort of 2691 individuals with neurodevelopmental conditions including ASD, schizophrenia, attention deficit hyperactivity disorder (ADHD), and obsessive–compulsive disorder20. Almost half of this total cohort (46%, 1230/2691) was assessed for intellectual disability (ID), of which 14.9% (183/1230) received the diagnosis and thus had ID combined with ASD, schizophrenia, or ADHD. We used CNVs consistently identified by multiple CNV calling algorithms (“stringent” CNVs) and were identified either as “rare” (< 0.1% in a control population) or being deemed of possible clinical relevance (see Table 2 in Zarrei et al.20). After removing CNVs from 17 individuals with aneuploidies, the final case data contained 1357 gain CNVs and 835 loss CNVs. We included inheritance and clinical classification data when available. Both cohorts were assessed using the Cytoscan HD microarray platform with the same CNV calling algorithms and similar sizing and probe cutoffs.
For each CNV, we paired every gene with its respective NeuroSCORE, then generated a median and average NeuroSCORE for the CNV. To simplify some of these analyses, we converted NeuroSCOREs to percentages of the total possible points (1 = 20%, 2 = 40%, 3 = 60%, 4 = 80%, 5 = 100%). We used two-sided t-tests to compare differences in CNVs between cases and controls for CNV size, total gene content, and the average and median NeuroSCORE. We then used Pearson’s chi squared or Fisher’s exact test (if N ≤ 5) to compare the distributions of the maximum scored gene within case and control CNVs. We performed sub-analyses using two-sided t-tests to explore differences in NeuroSCORE and scored genes by inheritance (inherited vs. de novo), gender (male vs. female proband), and clinical significance (common population variants, likely benign variants, variant of uncertain significance (VOUS), and likely clinically significant/clinically significant).
Finally, we performed linear regression analysis on classification and gene content by assigning increasing values to increasing pathogenicity and using the total count of genes in the CNV (see “Results”, Fig. 3 and case–control analyses). Zarrei et al.20 classified CNV pathogenicity, though we added a classification for common, “population variants” (CNVs observed at > 1% in the cohorts). Classification was coded as 1 = population variant, 2 = likely benign, 3 = VOUS, 4 = pathogenic/likely pathogenic.
NeuroSCORE in brain-expressed genes from the human protein atlas
Using the Human Protein Atlas, we analyzed genes defined as having “elevated” brain expression (N = 2587 genes), corresponding to gene expression within a brain region or cell type > 4 × higher than genes expressed in another tissue or cell type68. Using this set of genes, we compared the distribution of high scoring and any scoring genes to genome-wide NeuroSCORE data to determine if scored genes were enriched in this data (retrieved November 3, 2021 from https://www.proteinatlas.org/humanproteome/brain). After removing 145 genes due to not being a protein coding gene or a lack of scoring data, our data set included 2442 genes for analysis. We used chi squared tests to determine if the distribution of high scoring genes or any scored gene in the “elevated” brain expression data set was different than the distribution of these genes in the genome.
GO and pathway analyses
Gene ontology (GO) enrichment analysis was performed for the set of high scoring genes (identified by 3 or more gene metrics in our final model; N = 1601)69,70,71. We performed analyses for biological processes, molecular function, and cellular component using Bonferroni correction for multiple testing. We chose this correction as it is the most conservative correction. If multiple related terms were within the top enriched GO terms, we included the more specific term and omitted the broader term. This analysis used databased from the Gene Ontology Consortium (http://geneontology.org/) using the March 23rd, 2020 release.
To map the relevant pathways in which the high scoring genes were primarily involved, gene enrichment analysis was performed using the gene overlap package of R, followed by Cytoscape analysis to trace the pathways involved and their connectivity. The false decision rate and p-value cut off was 0.01 and 0.001, respectively. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) database were used for both gene enrichment and Cytoscape analysis. Then, the network was built using the enrichment map and the auto annotate Cytoscape application. The node color represents the p-value (the darker the shade, the lower the p-value) and size of the node represents increasing odds ratio.
Evaluation of NeuroSCORE model in mouse model data
We used the high-level phenotypic data provided by the Jackson Laboratory’s Mouse Genome Informatics database to further assess our model (accessed July 6th, 2020; http://www.informatics.jax.org)18. We downloaded all annotated genes and matched the mouse and human gene using the unique MGI number via HGNC database. We excluded genes where the human homolog of the mouse gene included two or more unique human genes (e.g., the mouse gene Rln1 is a homolog of both human RLN1 and RLN2). We also removed multiple mouse genes that matched the same human ortholog (e.g., mouse genes SCD1, SCD2, SCD3, and SCD4 are homologues of human SCD). Lastly, we removed genes where no mouse phenotype information was available, as a lack of high-level phenotype information does not mean variants of a gene cannot cause a phenotype. In total, we removed: 70 genes with multiple mouse homologues matching to a single human gene, 45 human genes that did not have a mouse ortholog, and 4139 genes without phenotype data. The total number of genes included for analysis with phenotype data was 8149, which included 3863 genes associated with any neurological phenotype and 4286 genes associated with any non-neurological phenotype. Of note, this data was not used to develop or refine the NeuroSCORE model.
Data availability
The data that support the findings of this study are available from Bionano Genomics, Inc., but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data, however, may be available from the authors upon reasonable request and with permission of Bionano Genomics, Inc.
References
Koch, T. 1831: The map that launched the idea of global health. Int. J. Epidemiol. 43, 1014–1020 (2014).
Tulchinsky, T. H. John Snow, Cholera, the broad street pump; waterborne diseases then and now. Case Stud. Public Heal. https://doi.org/10.1016/B978-0-12-804571-8.00017-2 (2018).
Srivastava, S. et al. Meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 21, 2413–2421 (2019).
Fitzgerald, T. et al. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015).
Bardakjian, T. M. et al. Genetic test utilization and diagnostic yield in adult patients with neurological disorders. Neurogenetics 19, 105–110 (2018).
Tammimies, K. et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. J. Am. Med. Assoc. 314, 895–903 (2015).
Heide, S. et al. Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: contribution to further diagnostic delineation. Genet. Med. 22, 1887–1891 (2020).
Papuc, S. M. et al. The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur. J. Hum. Genet. 27, 408–421 (2019).
Kaplanis, J. et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature 10, 757–762 (2020).
Pandey, A. K., Lu, L., Wang, X., Homayouni, R. & Williams, R. W. Functionally enigmatic genes: a case study of the brain ignorome. PLoS ONE 9, 1–11 (2014).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Havrilla, J. M., Pedersen, B. S., Layer, R. M. & Quinlan, A. R. A map of constrained coding regions in the human genome. Nat. Genet. 51, 88–95 (2019).
Karczewski, K. J. et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv https://doi.org/10.1101/531210 (2019).
Brueggeman, L., Koomar, T. & Michaelson, J. J. Forecasting risk gene discovery in autism with machine learning and genome-scale data. Sci. Rep. 10, 1–11 (2020).
Freytag, S., Burgess, R., Oliver, K. L. & Bahlo, M. brain-coX: Investigating and visualising gene co-expression in seven human brain transcriptomic datasets. Genome Med. 9, 1–9 (2017).
Moreno-De-Luca, A. et al. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 12, 406–414 (2013).
Gonzalez-Mantilla, A. J., Moreno-De-Luca, A., Ledbetter, D. H. & Martin, C. L. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psych. 73, 275–283 (2016).
Bult, C. J. et al. Mouse genome database (MGD) 2019. Nucl. Acids Res. 47, D801–D806 (2019).
Uddin, M. et al. A high-resolution copy number variation resource for clinical and population genetics. Genet. Med. 17, 747–752 (2015).
Zarrei, M. et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. Genomic Med. 4, (2019).
Jacquemont, S. et al. A higher mutational burden in females supports a ‘female protective model’ in neurodevelopmental disorders. Am. J. Hum. Genet. 94, 415–425 (2014).
Ho, K. S. et al. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders using an ultra-high resolution chromosomal microarray optimized for neurodevelopmental disorders. Int. J. Mol. Sci. 17, 1–14 (2016).
Kopp, N., McCullough, K., Maloney, S. E. & Dougherty, J. D. Gtf2i and Gtf2ird1 mutation do not account for the full phenotypic effect of the Williams syndrome critical region in mouse models. Hum. Mol. Genet. 28, 3443–3465 (2019).
Chailangkarn, T., Noree, C. & Muotri, A. R. The contribution of GTF2I haploinsufficiency to Williams syndrome. Mol. Cell. Probes 40, 45–51 (2018).
Cartier, E. et al. Rare autism-associated variants implicate syntaxin 1 (STX1 R26Q) phosphorylation and the dopamine transporter (hDAT R51W) in dopamine neurotransmission and behaviors. EBioMedicine 2, 135–146 (2015).
Reuter, M. S. et al. Diagnostic yield and novel candidate genes by exome sequencing in 152 consanguineous families with neurodevelopmental disorders. JAMA Psych. 74, 293–299 (2017).
Kosmicki, J. A. et al. Refining the role of de novo protein truncating variants in neurodevelopmental disorders using population reference samples. Nat. Genet. 49, 504–510 (2017).
Morris, C. A. et al. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am. J. Med. Genet. 123A, 45–59 (2003).
Mao, R. et al. LIMK1 and LIMK2 regulate cortical development through affecting neural progenitor cell proliferation and migration. Mol. Brain 12, 67 (2019).
Lalli, M. A. et al. Haploinsufficiency of BAZ1B contributes to Williams syndrome through transcriptional dysregulation of neurodevelopmental pathways. Hum. Mol. Genet. 25, 1294–1306 (2016).
Li, Z. et al. The mutational burden and oligogenic inheritance in klippel-feil syndrome. BMC Musculoskelet. Disord. 21, 1–9 (2020).
Capossela, S. et al. Growth defects and impaired cognitivebehavioral abilities in mice with knockout for Eif4h, a gene located in the mouse homolog of the Williams-beuren syndrome critical region. Am. J. Pathol. 180, 1121–1135 (2012).
Willsey, J. et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 13, 216–221 (2014).
Seidlitz, J. et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat. Commun. 11, 1–14 (2020).
Zhao, Y. et al. Variance of IQ is partially dependent on deletion type among 1427 22q11.2 deletion syndrome subjects. Am. J. Med. Genet. A 176, 2172–2181 (2018).
Lovrecic, L. et al. Microduplication in the 2p16.1p15 chromosomal region linked to developmental delay and intellectual disability. Mol. Cytogenet. 11, 1–6 (2018).
Mimouni-Bloch, A., Yeshaya, J., Kahana, S., Maya, I. & Basel-Vanagaite, L. A de-novo interstitial microduplication involving 2p16.1-p15 and mirroring 2p16.1-p15 microdeletion syndrome: clinical and molecular analysis. Eur. J. Paediatr. Neurol. 19, 711–715 (2015).
Chen, C.-P. et al. Prenatal diagnosis of a 3.2-Mb 2p16.1-p15 duplication associated with familial intellectual disability. Taiwan J. Obstet. Gynecol. 57, 578–582 (2018).
Bagheri, H. et al. Identifying candidate genes for 2p15p16.1 microdeletion syndrome using clinical, genomic, and functional analysis. JCI Insight 1, 1–20 (2016).
Wang, Y.-H., Ding, Z.-Y., Cheng, Y.-J., Chien, C.-T. & Huang, M.-L. An efficient screen for cell-intrinsic factors identifies the chaperonin CCT and multiple conserved mechanisms as mediating dendrite morphogenesis. Front. Cell. Neurosci. 14, 1–18 (2020).
Alfert, A., Moreno, N. & Kerl, K. The BAF complex in development and disease. Epigenet. Chromatin 12, 1–15 (2019).
Goodman, J. V. & Bonni, A. Regulation of neuronal connectivity in the mammalian brain by chromatin remodeling. Curr. Opin. Nuerobiol. 59, 59–68 (2019).
Van De Willige, D., Hoogenraad, C. C. & Akhmanova, A. Microtubule plus-end tracking proteins in neuronal development. Cell. Mol. Life Sci. 73, 2053–2077 (2016).
Mietelska-Porowska, A., Wasik, U., Goras, M., Filipek, A. & Niewiadomska, G. Tau protein modifications and interactions: their role in function and dysfunction. Int. J. Mol. Sci. 15, 4671–4713 (2014).
Shukla, S. & Tekwani, B. L. Histone deacetylases inhibitors in neurodegenerative diseases, neuroprotection and neuronal differentiation. Front. Pharmacol. 11, 1–20 (2020).
Sawyer, I. A., Sturgill, D., Sung, M., Hager, G. L. & Dundr, M. Cajal body function in genome organization and transcriptome diversity. BioEssays 38, 1197–1208 (2016).
Bergstrand, S. et al. Biallelic mutations in WRAP53 result in dysfunctional telomeres, Cajal bodies and DNA repair, thereby causing Hoyeraal–Hreidarsson syndrome. Cell Death Dis. 11, (2020).
Limanaqi, F. et al. Promiscuous roles of autophagy and proteasome in neurodegenerative proteinopathies. Int. J. Mol. Sci. 21, 1–31 (2020).
Olthof, A. M., Rasmussen, J. S., Campeau, P. M. & Kanadia, R. N. Disrupted minor intron splicing is prevalent in Mendelian disorders. Mol. Genet. Genomic Med. 8, 1–14 (2020).
Thelen, M. P. & Kye, M. J. The role of RNA binding proteins for local mRNA translation: implications in neurological disorders. Front. Mol. Biosci. 6, 1–13 (2020).
Rodriguez, J. M., Pozo, F., Di Domenico, T., Vazquez, J. & Tress, M. L. An analysis of tissue-specific alternative splicing at the protein level. PLoS Comput. Biol. 16, 1–24 (2020).
Garcia-Forn, M., Boitnott, A., Akpinar, Z. & De Rubeis, S. Linking autism risk genes to disruption of cortical development. Cells 9, 1–24 (2020).
Guillen Sacoto, M. J. et al. De Novo variants in the ATPase module of MORC2 cause a neurodevelopmental disorder with growth retardation and variable craniofacial dysmorphism. Am. J. Hum. Genet. 107, 352–363 (2020).
Williams, S. M. et al. An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol. Psych. 24, 1707–1719 (2019).
Martin, H. C. et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science 362, 1161–1164 (2018).
Coe, B. P. et al. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat. Genet. 51, 106–116 (2019).
Takata, A., Ionita-laza, I., Gogos, J. A., Xu, B. & Karayiorgou, M. D. Novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron 89, 940–947 (2016).
Martin, J. et al. A brief report: de novo copy number variants in children with attention deficit hyperactivity disorder. Transl. Psych. 10, 1–6 (2020).
Jin, S. C. et al. Mutations disrupting neuritogenesis genes confer risk for cerebral palsy. Nat. Genet. 52, 1046–1056 (2020).
Allen, A. S. et al. De novo mutations in the classic epileptic encephalopathies. Nature 18, 1067–1073 (2013).
Zou, F. et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 8, 1–16 (2012).
Coe, B. P. et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46, 1063–1071 (2014).
Uddin, M. et al. Indexing effects of copy number variation on genes involved in developmental delay. Sci. Rep. 6, 1–12 (2016).
Consortium & Gte,. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Turner, T. N. et al. denovo-db: a compendium of human de novo variants. Nucl. Acids Res. 45, D804–D811 (2017).
Uddin, M. et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat. Genet. 46, 742–747 (2014).
Ashwell, K. W. S. & Mai, J. K. Chapter 3-fetal development of the central nervous system. in The Human Nervous System (Third Edition) (eds. Mai, J. K. & Paxinos, G.) 31–79 (Academic Press, 2012). doi:https://doi.org/10.1016/B978-0-12-374236-0.10003-3
Sjöstedt, E., et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, (2020).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucl. Acids Res. 47, D419–D426 (2019).
Consortium, T. G. O., et al. Gene ontology : tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Consortium, T. G. O. The gene ontology resource: 20 years and still GOing strong. Nucl. Acids Res. 47, 330–338 (2019).
Acknowledgements
We would like to thank Dr. Lori Erby, ScM, CGC and Colleen Caleshu, ScM, CGC for helpful comments and thoughts on this project, as well as Dr. Aric Schadler for support with statistical methodology.
Author information
Authors and Affiliations
Contributions
Study conception and design: K.W.D., C.G.B., M.M., R.V., M.S.; Data collection: K.W.D., M.R.; Statistical Analyses: K.W.D., M.R.; Model application analyses: K.W.D., C.G.B., Y.H., M.U., M.S.; Data interpretation: K.W.D., C.G.B., M.M., Y.H., M.U., M.S.; Writing and editing: K.W.D., C.G.B., M.M., R.V., M.U., M.S.
Corresponding author
Ethics declarations
Competing interests
Megan Martin, Rena Vanzo, Dr. Colleen G. Bilancia, and Dr. Moises Serrano all are employees of and hold stock options in Bionano Genomics, Inc. Kyle W. Davis and Megan Rimmasch were previously employed by Bionano Genomics, Inc. and Kyle W. Davis own stock options in Bionano Genomics, Inc. Dr. Mohammed Uddin is listed as an inventor on a patent application for his work on critical exon indexing (upon which the Index gene metric is based); the patent is held by Toronto Hospital for Sick Children and licensed by Lineagen Inc, a Bionano Genomics Company. Yolanda Hom has declared no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Davis, K.W., Bilancia, C.G., Martin, M. et al. NeuroSCORE is a genome-wide omics-based model that identifies candidate disease genes of the central nervous system. Sci Rep 12, 5427 (2022). https://doi.org/10.1038/s41598-022-08938-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-08938-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.