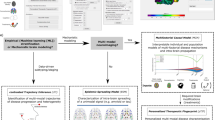
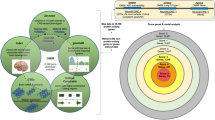
Abstract
A major challenge in the treatment of neurodegenerative disorders is the translation of effective therapies from the lab to the clinic. One approach to improve this process is the use of human brain tissue microarray (HBTMA) technology to aid in the discovery and validation of drug targets for brain disorders. In this protocol we describe a platform for the production of high-quality HBTMAs that can be used for drug target discovery and validation. We provide examples of the use of this platform and describe detailed protocols for HBTMA design, construction and use for both protein and mRNA detection. This platform requires less tissue and reagents than single-slide approaches, greatly increasing throughput and capacity, enabling samples to be compared in a more consistent way. It takes 4 d to construct a 60 core HBTMA. Immunohistochemistry and in situ hybridization take a further 2 d. Imaging of each HBTMA slide takes 15 min, with subsequent high-content analysis taking 30 min–2 h.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Datasets generated and described in our supporting primary papers can be made available from the corresponding author upon reasonable request. An example of data generated using this protocol is available in Supplementary Table 1.
References
Dragunow, M. Human brain neuropharmacology: a platform for translational neuroscience. Trends Pharmacol. Sci. 41, 777–792 (2020).
Singh-Bains, M. K. et al. Altered microglia and neurovasculature in the Alzheimer’s disease cerebellum. Neurobiol. Dis. 132, 104589 (2019).
Coppieters, N. et al. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol. Aging 35, 1334–1344 (2014).
Narayan, P. J. et al. Assessing fibrinogen extravasation into Alzheimer’s disease brain using high-content screening of brain tissue microarrays. J. Neurosci. Methods 247, 41–49 (2015).
Narayan, P. J., Lill, C., Faull, R., Curtis, M. A. & Dragunow, M. Increased acetyl and total histone levels in post-mortem Alzheimer’s disease brain. Neurobiol. Dis. 74, 281–294 (2015).
Dominy, S. S. et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5, eaau3333 (2019).
Singh‐Bains, M. K. et al. Globus pallidus degeneration and clinico‐pathological features of Huntington’s disease. Ann. Neurol. 80, 185–201 (2016).
Singh‐Bains, M. K. et al. Cerebellar degeneration correlates with motor symptoms in Huntington disease. Ann. Neurol. 85, 396–405 (2019).
Mehrabi, N. F. et al. Symptom heterogeneity in Huntington’s disease correlates with neuronal degeneration in the cerebral cortex. Neurobiol. Dis. 96, 67–74 (2016).
Kim, E. H. et al. Cortical interneuron loss and symptom heterogeneity in Huntington disease. Ann. Neurol. 75, 717–727 (2014).
Thu, D. C. et al. Cell loss in the motor and cingulate cortex correlates with symptomatology in Huntington’s disease. Brain 133, 1094–1110 (2010).
Nana, A. L. et al. Widespread heterogeneous neuronal loss across the cerebral cortex in Huntington’s disease. J. Huntingtons Dis. 3, 45–64 (2014).
West, M. J. Design-based stereological methods for counting neurons. Prog. Brain Res. 135, 43–51 (2002).
Kononen, J. et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4, 844–847 (1998).
Alkushi, A. et al. Immunoprofile of cervical and endometrial adenocarcinomas using a tissue microarray. Virchows Arch. 442, 271–277 (2003).
Garcıa, J. F. et al. Hodgkin and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue microarrays. Blood 101, 681–689 (2003).
Makretsov, N. A. et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clin. Cancer Res. 10, 6143–6151 (2004).
Giltnane, J. M. & Rimm, D. L. Technology insight: identification of biomarkers with tissue microarray technology. Nat. Clin. Pract. Oncol. 1, 104–111 (2004).
Camp, R. L., Dolled-Filhart, M., King, B. L. & Rimm, D. L. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 63, 1445–1448 (2003).
Martikainen, P., Louhelainen, A.-M., Kauppinen, T. & Alafuzoff, I. Human brain tissue microarrays as a platform to investigate diseases of the nervous system. Brain Res. 1089, 33–43 (2006).
Dancau, A.-M., Simon, R., Mirlacher, M. & Sauter, G. in Cancer Gene Profiling 53–65 (Springer, 2016).
Yang, P., Waldvogel, H., Turner, C., Faull, R. & Dragunow, M. Vascular remodelling is impaired in Parkinson disease. J. Alzheimers Dis. Parkinsonism 7, 2161–0460.1000313 (2017).
Hewitt, S. M. in Molecular Profiling 201–214 (Springer, 2012).
Eguiluz, C., Viguera, E., Millán, L. & Pérez, J. Multitissue array review: a chronological description of tissue array techniques, applications and procedures. Pathol. Res. Pract. 202, 561–568 (2006).
Tzankov, A., Went, P., Zimpfer, A. & Dirnhofer, S. Tissue microarray technology: principles, pitfalls and perspectives—lessons learned from hematological malignancies. Exp. Gerontol. 40, 737–744 (2005).
Hassan, S., Ferrario, C., Mamo, A. & Basik, M. Tissue microarrays: emerging standard for biomarker validation. Curr. Opin. Biotechnol. 19, 19–25 (2008).
Packeisen, J., Korsching, E., Herbst, H., Boecker, W. & Buerger, H. Demystified…tissue microarray technology. Mol. Pathol. 56, 198 (2003).
Simon, R. in Tissue Microarrays 1–16 (Springer, 2010).
Hutchins, G. & Grabsch, H. I. How to make tissue microarrays. Diagn. Histopathol. 24, 127–135 (2018).
Nedjadi, T. et al. Prognostic value of HER2 status in bladder transitional cell carcinoma revealed by both IHC and BDISH techniques. BMC Cancer 16, 653 (2016).
Plum, P. S. et al. HER2/neu (ERBB2) expression and gene amplification correlates with better survival in esophageal adenocarcinoma. BMC Cancer 19, 38 (2019).
Parsons, M. & Grabsch, H. How to make tissue microarrays. Diagn. Histopathol. 15, 142–150 (2009).
Gulmann, C. & O’Grady, A. Tissue microarrays: an overview. Curr. Diagn. Pathol. 9, 149–154 (2003).
Dragunow, M. The adult human brain in preclinical drug development. Nat. Rev. Drug Discov. 7, 659–666 (2008).
Kauppinen, T., Martikainen, P. & Alafuzoff, I. Human postmortem brain tissue and 2-mm tissue microarrays. Appl. Immunohistochem. Mol. Morphol. 14, 353–359 (2006).
Sjöbeck, M., Haglund, M., Persson, A., Sturesson, K. & Englund, E. Brain tissue microarrays in dementia research: white matter microvascular pathology in Alzheimer’s disease. Neuropathology 23, 290–295 (2003).
Wang, H., Wang, H., Zhang, W. & Fuller, G. N. Tissue microarrays: applications in neuropathology research, diagnosis, and education. Brain Pathol. 12, 95–107 (2002).
Goldstine, J., Seligson, D. B., Beizai, P., Miyata, H. & Vinters, H. V. Tissue microarrays in the study of non-neoplastic disease of the nervous system. J. Neuropathol. Exp. Neurol. 61, 653–662 (2002).
Walker, L. et al. Quantitative neuropathology: an update on automated methodologies and implications for large scale cohorts. J. Neural Transm. 124, 671–683 (2017).
Eckel-Passow, J. E. et al. Tissue microarrays: one size does not fit all. Diagn. Pathol. 5, 48 (2010).
Dhir, R. in Tissue Proteomics (eds Liu, B. C. S. & Ehrlich, J. R.) 91–103 (Humana Press, 2008).
Gately, K., Kerr, K. & O’Byrne, K. in Gene Expression Profiling 139–153 (Springer, 2011).
Hegde, R. N. et al. TBK1 phosphorylates mutant Huntingtin and suppresses its aggregation and toxicity in Huntington’s disease models. EMBO J. 39, e104671 (2020).
Waldvogel, H. et al. The collection and processing of human brain tissue for research. Cell Tissue Bank. 9, 169–179 (2008).
Waldvogel, H. J., Curtis, M. A., Baer, K., Rees, M. I. & Faull, R. L. M. Immunohistochemical staining of post-mortem adult human brain sections. Nat. Protoc. 1, 2719–2732 (2006).
Howat, W. J. & Wilson, S. J. in Tissue Microarrays 63–72 (Springer, 2010).
Mirra, S. S. et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD): Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–479 (1991).
Braak, H. & Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 (1991).
Wan, W.-H., Fortuna, M. B. & Furmanski, P. A rapid and efficient method for testing immunohistochemical reactivity of monoclonal antibodies against multiple tissue samples simultaneously. J. Immunol. Methods 103, 121–129 (1987).
Vogel, U. F. The construction of high-density paraffin tissue microarrays with 0.43-mm-diameter paraffin tissue core biopsies is technically feasible. Virchows Arch. 453, 43–46 (2008).
Narayan, P. et al. Inconsistencies in histone acetylation patterns among different HD model systems and HD post-mortem brains. Neurobiol. Dis. 146, 105092 (2020).
Bolton, K. L. et al. Assessment of automated image analysis of breast cancer tissue microarrays for epidemiologic studies. Cancer Epidemiol. Biomark. Prev. 19, 992–999 (2010).
Braun, M. et al. Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol. Histopathol. 28, 605–610 (2013).
Camp, R. L., Chung, G. G. & Rimm, D. L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 8, 1323–1328 (2002).
Dolled-Filhart, M. et al. in Tissue Microarrays 151–162 (Springer, 2010).
Ryan, D., Mulrane, L., Rexhepaj, E. & Gallagher, W. M. in Drug Safety Evaluation 97–112 (Springer, 2011).
Loughrey, M. B. et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology 73, 327–338 (2018).
Dragunow, M. High-content analysis in neuroscience. Nat. Rev. Neurosci. 9, 779–788 (2008).
Wang, F. et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012).
Acknowledgements
We express our appreciation to all donor families in New Zealand, who through their generosity have enabled their invaluable tissue donation to the Neurological Foundation Human Brain Bank for the construction of HBTMAs. We thank M. A. Curtis, Deputy Director of the Neurological Foundation Human Brain Bank. We thank Senior Histologist S. Amirapu for her contribution to tissue processing and paraffin embedding protocols for TMA preparation. We thank C. Turner for his expert neuropathological assessments of the cases utilized for TMA construction. We acknowledge the excellent work and assistance of M. Eszes (Human Brain Bank), K. Hubbard and C. Lill (Research Technicians). We acknowledge the staff at the Biomedical Imaging Research Unit (R. Kurian and P. V. Anekal) for VSlide Scanner support. We acknowledge P. J. Narayan and E. L. Scotter for their contributions to the development of the TMA imaging and analysis protocols (references cited in the manuscript). We acknowledge M. D. R. Austria for his contribution to the presubmission enquiry. This work was supported by a Programme Grant from the Health Research Council of New Zealand, Brain Research New Zealand, Neurological Foundation of New Zealand, the Hugh Green Foundation, The Coker Trust, the Sir Thomas and Lady Duncan Trust and the Freemasons Foundation of New Zealand. MKSB was supported by the Leo Nilon Huntington’s Disease Research Fellowship.
Author information
Authors and Affiliations
Contributions
M.K.S.B. and N.F.M. contributed equally to manuscript production, figure production, HBTMA design, construction and analysis protocols. A.Y.S.T. contributed to figure production and HBTMA analysis protocols. R.L.M.F. supervised the ethical collection, donor interaction, clinical assessment and processing of human brain tissue for the HBTMA platform. M.D. conceived, established and directs the HBTMA platform and high-content-analysis platform. M.K.S.B. and N.F.M. wrote the draft and completed the final manuscript with the contributions, edits and approvals of the final manuscript from A.Y.S.T., R.L.M.F. and M.D.
Corresponding author
Ethics declarations
Competing interests
M.D. and R.L.M.F. have developed a platform called Neurovalida for commercial use of human brain tissue microarray.
Additional information
Peer review information Nature Protocols thanks Christel Herold-Mende and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Dominy, S. S. et al. Sci. Adv. 5, eaau3333 (2019): https://doi.org/10.1126/sciadv.aau3333
Singh-Bains, M. K. et al. Neurobiol. Dis. 132, 104589 (2019): https://doi.org/10.1016/j.nbd.2019.104589
Narayan, P. J. et al. J. Neurosci. Methods 247, 41–49 (2015): https://doi.org/10.1016/j.jneumeth.2015.03.017
Supplementary information
Supplementary Table 1
Supplementary Table 1 contains example data outputs generated from count nuclei (gray heading), neurite outgrowth (pink heading) and optical density scaled (yellow heading) Metamorph journals
Rights and permissions
About this article
Cite this article
Singh-Bains, M.K., Mehrabi, N.F., Tan, A.Y.S. et al. Preparation, construction and high-throughput automated analysis of human brain tissue microarrays for neurodegenerative disease drug development. Nat Protoc 16, 2308–2343 (2021). https://doi.org/10.1038/s41596-021-00503-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00503-7
This article is cited by
-
Aggregate-prone brain regions in Parkinson’s disease are rich in unique N-terminus α-synuclein conformers with high proteolysis susceptibility
npj Parkinson's Disease (2024)
-
Neutrophil-vascular interactions drive myeloperoxidase accumulation in the brain in Alzheimer’s disease
Acta Neuropathologica Communications (2022)
-
Characterisation of PDGF-BB:PDGFRβ signalling pathways in human brain pericytes: evidence of disruption in Alzheimer’s disease
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.