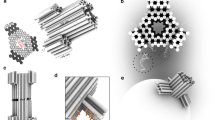
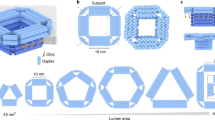
Abstract
DNA nanopores are bio-inspired nanostructures that control molecular transport across lipid bilayer membranes. Researchers can readily engineer the structure and function of DNA nanopores to synergistically combine the strengths of DNA nanotechnology and nanopores. The pores can be harnessed in a wide range of areas, including biosensing, single-molecule chemistry, and single-molecule biophysics, as well as in cell biology and synthetic biology. Here, we provide a protocol for the rational design of nanobarrel-like DNA pores and larger DNA origami nanopores for targeted applications. We discuss strategies for the pores’ chemical modification with lipid anchors to enable them to be inserted into membranes such as small unilamellar vesicles (SUVs) and planar lipid bilayers. The procedure covers the self-assembly of DNA nanopores via thermal annealing, their characterization using gel electrophoresis, purification, and direct visualization with transmission electron microscopy and atomic force microscopy. We also describe a gel assay to determine pore–membrane binding and discuss how to use single-channel current recordings and dye flux assays to confirm transport through the pores. We expect this protocol to take approximately 1 week to complete for DNA nanobarrel pores and 2–3 weeks for DNA origami pores.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout













Similar content being viewed by others
Data availability
All data are available from the corresponding author upon reasonable request.
References
Tunuguntla, R. H., Allen, F. I., Kim, K., Belliveau, A. & Noy, A. Ultrafast proton transport in sub-1-nm diameter carbon nanotube porins. Nat. Nanotechnol. 11, 639–644 (2016).
Feng, J. et al. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotechnol. 10, 1070–1076 (2015).
Yusko, E. C. et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 12, 360–367 (2017).
Huang, G., Voet, A. & Maglia, G. FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 Dalton resolution. Nat. Commun. 10, 835 (2019).
Wei, R. S., Gatterdam, V., Wieneke, R., Tampe, R. & Rant, U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 7, 257–263 (2012).
Thakur, A. K. & Movileanu, L. Real-time measurement of protein-protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 37, 96–101 (2019).
Pulcu, G. S., Mikhailova, E., Choi, L. S. & Bayley, H. Continuous observation of the stochastic motion of an individual small-molecule walker. Nat. Nanotechnol. 10, 76–83 (2015).
Miles, B. N. et al. Single molecule sensing with solid-state nanopores: novel materials, methods, and applications. Chem. Soc. Rev. 42, 15–28 (2013).
Varongchayakul, N., Song, J., Meller, A. & Grinstaff, M. W. Single-molecule protein sensing in a nanopore: a tutorial. Chem. Soc. Rev. 47, 8512–8524 (2018).
Restrepo-Pérez, L., Joo, C. & Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 13, 786–796 (2018).
Alberts, B. et al. Molecular Biology of the Cell 6th edn (Garland Publishing, 2014).
Quick, J. et al. Real-time, portable genome sequencing for ebola surveillance. Nature 530, 228–232 (2016).
Clarke, J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009).
Cherf, G. M. et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-Å precision. Nat. Biotechnol. 30, 344–348 (2012).
Manrao, E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012).
Majd, S. et al. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 21, 439–476 (2010).
Galenkamp, N. S., Soskine, M., Hermans, J., Wloka, C. & Maglia, G. Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores. Nat. Commun. 9, 4085 (2018).
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Schoch, R. B., Han, J. & Renaud, P. Transport phenomena in nanofluidics. Rev. Mod. Phys. 80, 839 (2008).
Mahendran, K. R. et al. A monodisperse transmembrane α-helical peptide barrel. Nat. Chem. 9, 411–419 (2017).
Gopfrich, K. et al. Ion channels made from a single membrane-spanning DNA duplex. Nano Lett. 16, 4665–4669 (2016).
Gopfrich, K. et al. DNA-tile structures induce ionic currents through lipid membranes. Nano Lett. 15, 3134–3138 (2015).
Langecker, M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012).
Burns, J. R., Seifert, A., Fertig, N. & Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 11, 152–156 (2016).
Burns, J., Stulz, E. & Howorka, S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 13, 2351–2356 (2013).
Seifert, A. et al. Bilayer-spanning DNA nanopores with voltage-switching between open and closed state. ACS Nano 9, 1117–1126 (2015).
Arnott, P. M., Joshi, H., Aksimentiev, A. & Howorka, S. Dynamic interactions between lipid-tethered DNA and phospholipid membranes. Langmuir 34, 15084–15092 (2018).
Chen, L., Liang, S., Chen, Y., Wu, M. & Zhang, Y. Destructing the plasma membrane with activatable vesicular DNA nanopores. ACS Appl. Mater. Interfaces 12, 96–105 (2020).
Peng, R. et al. DNA-based artificial molecular signaling system that mimics basic elements of reception and response. Nat. Commun. 11, 978 (2020).
Gopfrich, K. et al. Large-conductance transmembrane porin made from DNA origami. ACS Nano 10, 8207–8214 (2016).
Krishnan, S. et al. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 7, 12787 (2016).
Diederichs, T. et al. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 10, 5018 (2019).
Thomsen, R. P. et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 10, 5655 (2019).
Chidchob, P. et al. Spatial presentation of cholesterol units on a DNA cube as a determinant of membrane protein-mimicking functions. J. Am. Chem. Soc. 141, 1100–1108 (2019).
Li, C. et al. Single-stranded DNA designed lipophilic G-quadruplexes as transmembrane channels for switchable potassium transport. Chem. Commun. 55, 12004–12007 (2019).
Burns, J. R. et al. Lipid bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angew. Chem. Int. Ed. 52, 12069–12072 (2013).
Burns, J. R., Al-Juffali, N., Janes, S. M. & Howorka, S. Membrane-spanning DNA nanopores with cytotoxic effect. Angew. Chem. Int. Ed. 53, 12466–12470 (2014).
Messager, L. et al. Biomimetic hybrid nanocontainers of designed permeability. Angew. Chem. Int. Ed. 55, 11106–11109 (2016).
Arnott, P. M. & Howorka, S. A temperature-gated nanovalve self-assembled from DNA to control molecular transport across membranes. ACS Nano 13, 3334–3340 (2019).
Kuzuya, A., Wang, R., Sha, R. & Seeman, N. C. Six-helix and eight-helix DNA nanotubes assembled from half-tubes. Nano Lett. 7, 1757–1763 (2007).
Mathieu, F. et al. Six-helix bundles designed from DNA. Nano Lett. 5, 661–665 (2005).
Langecker, M., Arnaut, V., List, J. & Simmel, F. C. DNA nanostructures interacting with lipid bilayer membranes. Acc. Chem. Res. 47, 1807–1815 (2014).
Ohmann, A. et al. Controlling aggregation of cholesterol-modified DNA nanostructures. Nucleic Acids Res. 47, 11441–11451 (2019).
Sacca, B. & Niemeyer, C. M. DNA origami: the art of folding DNA. Angew. Chem. Int. Ed. 51, 58–66 (2012).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Hong, F., Zhang, F., Liu, Y. & Yan, H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 117, 12584–12640 (2017).
Praetorius, F. et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017).
Pugh, G. C., Burns, J. R. & Howorka, S. Comparing proteins and nucleic acids for next-generation biomolecular engineering. Nat. Rev. Chem. 2, 113–130 (2018).
Litvinchuk, S. et al. Synthetic pores with reactive signal amplifiers as artificial tongues. Nat. Mater. 6, 576–580 (2007).
Sprengel, A. et al. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat. Commun. 8, 14472 (2017).
Bae, W., Kocabey, S. & Liedl, T. DNA nanostructures in vitro, in vivo and on membranes. Nano Today 26, 98–107 (2019).
Burns, J. R. & Howorka, S. Structural and functional stability of DNA nanopores in biological media. Nanomaterials 9, 490 (2019).
Ducani, C., Kaul, C., Moche, M., Shih, W. M. & Hogberg, B. Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides. Nat. Methods 10, 647–652 (2013).
Zhang, Z., Yang, Y., Pincet, F., C. L., M. & Lin, C. Placing and shaping liposomes with reconfigurable DNA nanocages. Nat. Chem. 9, 653–659 (2017).
Kuzyk, A. et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 483, 311–314 (2012).
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Zhang, Y. W. & Seeman, N. C. Construction of a DNA-truncated octahedron. J. Am. Chem. Soc. 116, 1661–1669 (1994).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Ohmann, A. et al. A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nat. Commun. 9, 2426 (2018).
Gut, I. G. & Beck, S. A procedure for selective DNA alkylation and detection by mass-spectrometry. Nucleic Acids Res. 23, 1367–1373 (1995).
Maingi, V. et al. Stability and dynamics of membrane-spanning DNA nanopores. Nat. Commun. 8, 14784 (2017).
Liu, P. et al. Charge neutralization drives the shape reconfiguration of DNA nanotubes. Angew. Chem. Int. Ed. 57, 5418–5422 (2018).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Kim, D. N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012).
Bellot, G., McClintock, M. A., Lin, C. & Shih, W. M. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat. Methods 8, 192–194 (2011).
Stahl, E., Martin, T. G., Praetorius, F. & Dietz, H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. 53, 12735–12740 (2014).
Xing, S. et al. Constructing higher-order DNA nanoarchitectures with highly purified DNA nanocages. ACS Appl. Mater. Interfaces 7, 13174–13179 (2015).
Shaw, A., Benson, E. & Hogberg, B. Purification of functionalized DNA origami nanostructures. ACS Nano 9, 4968–4975 (2015).
Bai, X. C., Martin, T. G., Scheres, S. H. & Dietz, H. Cryo-EM structure of a 3D DNA-origami object. Proc. Natl Acad. Sci. USA 109, 20012–20017 (2012).
Schreiber, R. et al. Hierarchical assembly of metal nanoparticles, quantum dots and organic dyes using DNA origami scaffolds. Nat. Nanotechnol. 9, 74–78 (2014).
Endo, M. & Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 47, 1645–1653 (2014).
Andersen, E. S. et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–76 (2009).
Wei, B., Dai, M. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Beckwitt, E. C., Kong, M. & Van Houten, B. Studying protein-DNA interactions using atomic force microscopy. Semin. Cell Dev. Biol. 73, 220–230 (2018).
Raghavan, G., Hidaka, K., Sugiyama, H. & Endo, M. Direct observation and analysis of the dynamics of the photoresponsive transcription factor GAL4. Angew. Chem. Int. Ed. 58, 7626–7630 (2019).
Franquelim, H. G., Khmelinskaia, A., Sobczak, J. P., Dietz, H. & Schwille, P. Membrane sculpting by curved DNA origami scaffolds. Nat. Commun. 9, 811 (2018).
Suzuki, Y., Endo, M. & Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nat. Commun. 6, 8052 (2015).
Hansma, H. G. & Laney, D. E. DNA binding to mica correlates with cationic radius: assay by atomic force microscopy. Biophys. J. 70, 1933–1939 (1996).
Mitchell, N., Ebner, A., Hinterdorfer, P., Tampe, R. & Howorka, S. Chemical tags mediate the orthogonal self-assembly of DNA duplexes into supramolecular structures. Small 6, 1732–1735 (2010).
Pyne, A., Thompson, R., Leung, C., Roy, D. & Hoogenboom, B. W. Single-molecule reconstruction of oligonucleotide secondary structure by atomic force microscopy. Small 10, 3257–3261 (2014).
Lyubchenko, Y. L., Shlyakhtenko, L. S. & Gall, A. A. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol. Biol. 543, 337–351 (2009).
Goyal, P. et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253 (2014).
Kučerka, N., Nieh, M. P. & Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochem. Biphys. Acta 1808, 2761–2771 (2011).
Burns, J. R. & Howorka, S. Defined bilayer interactions of DNA nanopores revealed with a nuclease-based nanoprobe strategy. ACS Nano 12, 3263–3271 (2018).
Birkholz, O. et al. Multi-functional DNA nanostructures that puncture and remodel lipid membranes into hybrid materials. Nat. Commun. 9, 1521 (2018).
Fogarty, J. C., Arjunwadkar, M., Pandit, S. A. & Pan, J. Atomically detailed lipid bilayer models for the interpretation of small angle neutron and x-ray scattering data. Biochim. Biophys. Acta 1848, 662–672 (2015).
Praetorius, F. & Dietz, H. Self-assembly of genetically encoded DNA-protein hybrid nanoscale shapes. Science 355, eaam5488 (2017).
Baaken, G., Ankri, N., Schuler, A. K., Ruhe, J. & Behrends, J. C. Nanopore-based single-molecule mass spectrometry on a lipid membrane microarray. ACS Nano 5, 8080–8088 (2011).
Jun, H. et al. Autonomously designed free-form 2D DNA origami. Sci. Adv. 5, eaav0655 (2019).
de Llano, E. et al. Adenita: interactive 3D modeling and visualization of DNA nanostructures. Nucleic Acids Res. 48, 8269–8275 (2020).
Zhang, F. et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 10, 779–784 (2015).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Lacroix, A., Edwardson, T. G. W., Hancock, M. A., Dore, M. D. & Sleiman, H. F. Development of DNA nanostructures for high-affinity binding to human serum albumin. J. Am. Chem. Soc. 139, 7355–7362 (2017).
Plesa, C. et al. Fast translocation of proteins through solid state nanopores. Nano Lett. 13, 658–663 (2013).
Acknowledgements
The authors thank J. Ciccone, R. Dickman, and H. Philpott for helping with figures and providing valuable feedback. Furthermore, the authors thank C. Weichbrodt from Nanion Technologies for providing feedback on the section of single-channel current recordings. The Howorka Group receives funding from the EPSRC (EP/ N009282/1), the BBSRC (BB/M025373/1, BB/N017331/1), and the Leverhulme Trust (RPG-2017-015). C.L. and E.G. are supported by the Biotechnology and Biological Sciences Research Council (BB/MO09513/1). C.L. is also supported by the National Physical Laboratory.
Author information
Authors and Affiliations
Contributions
C.L., D.O.-S., G.P., and J.R.B. provided the protocols for design and assembly. D.O.-S. and G.P. supplied the protocol for CanDo. C.L., D.O.-S, Y.X., and J.R.B. prepared the protocol for cholesterol lipid anchors. J.R.B. contributed the protocol for the alkyl modification of DNA. C.L. and D.O.-S. generated the protocol for gel electrophoretic characterization. E.G. prepared the protocols for purification of DNA origami structures. Y.X. provided the protocols for TEM analysis, and J.R.B. provided the protocols for AFM. C.L. provided the protocol for gel-binding assays. A.D. wrote the protocol for single-channel current recordings. C.L. provided the protocol for dye flux assays. D.O.-S. compiled the list of and set up materials and equipment. C.L., A.D., and J.R.B. supplied the troubleshooting advice. C.L., D.O.-S., J.R.B., and S.H. wrote the manuscript with input from all authors. C.L., D.O.-S, J.R.B., and S.H. edited the manuscript. C.L., A.D., G.P., and J.R.B. generated the figures, and J.R.B. edited them. All authors were part of the data analysis and discussions.
Corresponding authors
Ethics declarations
Competing interests
G.P., J.R.B., and S.H. hold patents on DNA nanopores that have been licensed to Oxford Nanopore Technologies.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Diederichs, T. et al. Nat. Comm. 10, 5018 (2019): https://doi.org/10.1038/s41467-019-12639-y
Burns, J. R., Seifert, A., Fertig, N. & Howorka, S. Nat. Nanotechnol. 11, 152–156 (2016): https://doi.org/10.1038/nnano.2015.279
Langecker, M. Science 338, 932–936 (2012): https://doi.org/10.1126/science.1225624
Burns, J. R., Stulz, E. & Howorka, S. Nano Lett. 13, 2351–2356 (2013): https://doi.org/10.1021/nl304147f
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Rights and permissions
About this article
Cite this article
Lanphere, C., Offenbartl-Stiegert, D., Dorey, A. et al. Design, assembly, and characterization of membrane-spanning DNA nanopores. Nat Protoc 16, 86–130 (2021). https://doi.org/10.1038/s41596-020-0331-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0331-7
This article is cited by
-
DNA-empowered synthetic cells as minimalistic life forms
Nature Reviews Chemistry (2024)
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
A lumen-tunable triangular DNA nanopore for molecular sensing and cross-membrane transport
Nature Communications (2024)
-
Functionalization and higher-order organization of liposomes with DNA nanostructures
Nature Communications (2023)
-
Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.